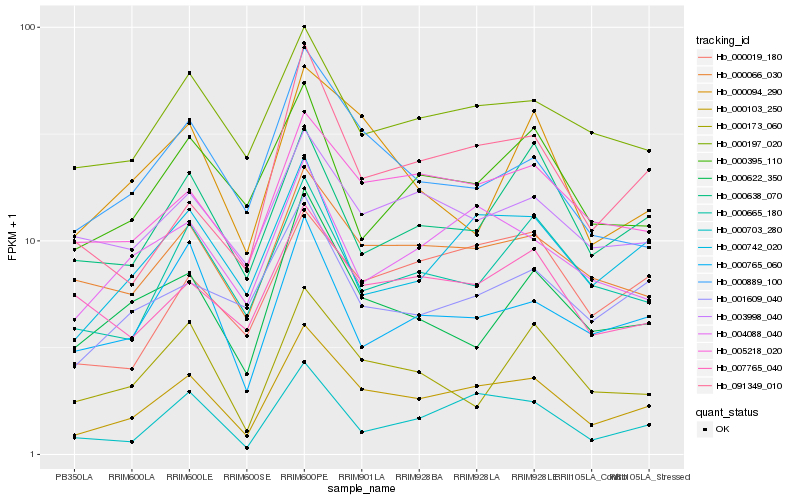
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_250 |
0.0 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 2 |
Hb_000066_030 |
0.1249711119 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000703_280 |
0.1271063183 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 4 |
Hb_005218_020 |
0.1280413088 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 5 |
Hb_000742_020 |
0.1305633211 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 6 |
Hb_000889_100 |
0.1310369993 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 7 |
Hb_000665_180 |
0.1359344343 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_000622_350 |
0.1362228782 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 9 |
Hb_091349_010 |
0.1364055388 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 10 |
Hb_000197_020 |
0.1368026171 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 11 |
Hb_004088_040 |
0.1378160724 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 12 |
Hb_000765_060 |
0.140737107 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 13 |
Hb_001609_040 |
0.1425793109 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 14 |
Hb_000019_180 |
0.1438527713 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 15 |
Hb_003998_040 |
0.146980103 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 16 |
Hb_000173_060 |
0.147124063 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000094_290 |
0.1500181829 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_007765_040 |
0.1502254871 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 19 |
Hb_000638_070 |
0.150501063 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 20 |
Hb_000395_110 |
0.1505161163 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |