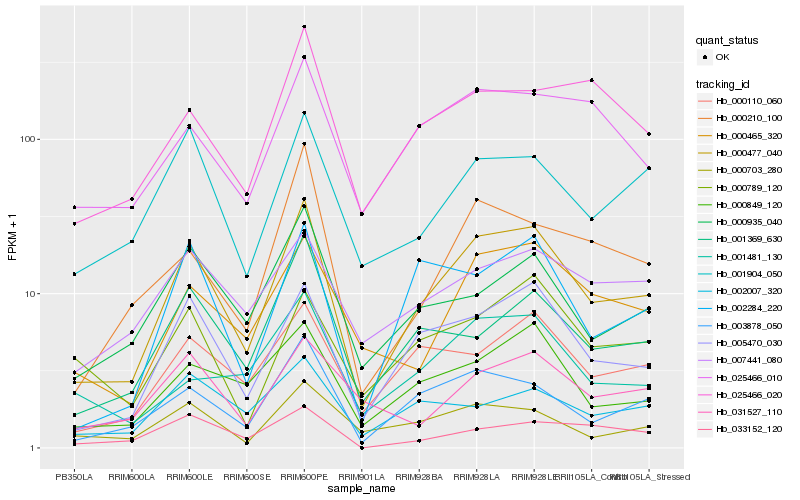
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005470_030 |
0.1157963077 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003878_050 |
0.1342173587 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 4 |
Hb_000849_120 |
0.1524947291 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 5 |
Hb_000110_060 |
0.1548032749 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 6 |
Hb_000465_320 |
0.1561272155 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 7 |
Hb_001904_050 |
0.1573379684 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 8 |
Hb_002284_220 |
0.1601273459 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 9 |
Hb_001369_630 |
0.1602644336 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 10 |
Hb_000935_040 |
0.160371164 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_000703_280 |
0.1690498073 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 12 |
Hb_000210_100 |
0.1717740962 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 13 |
Hb_025466_010 |
0.174574432 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_033152_120 |
0.1766626029 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 15 |
Hb_007441_080 |
0.1775248885 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 16 |
Hb_025466_020 |
0.1776927103 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 17 |
Hb_031527_110 |
0.1790402979 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 18 |
Hb_002007_320 |
0.1799998525 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 19 |
Hb_001481_130 |
0.1802767077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000789_120 |
0.1811880713 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |