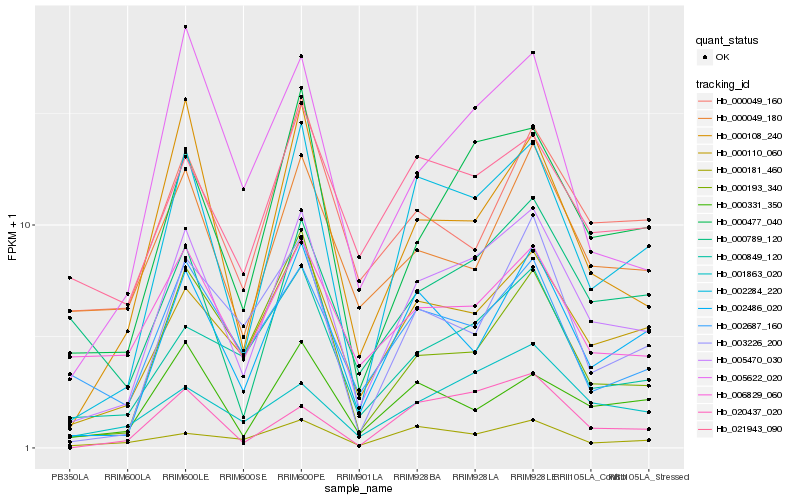
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005470_030 |
0.0 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002284_220 |
0.0959014346 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 3 |
Hb_000110_060 |
0.1150729221 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 4 |
Hb_000477_040 |
0.1157963077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002486_020 |
0.1356953019 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000849_120 |
0.1434115024 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 7 |
Hb_020437_020 |
0.1494135003 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 8 |
Hb_000108_240 |
0.1494944726 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 9 |
Hb_005622_020 |
0.158143185 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003226_200 |
0.1595137268 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 11 |
Hb_021943_090 |
0.1603877526 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 12 |
Hb_002687_160 |
0.161484748 |
- |
- |
PREDICTED: uncharacterized protein LOC105643395 [Jatropha curcas] |
| 13 |
Hb_001863_020 |
0.1618443093 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000789_120 |
0.1652427186 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000049_180 |
0.1671460068 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 16 |
Hb_000331_350 |
0.1683403794 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 17 |
Hb_000193_340 |
0.1693748854 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006829_060 |
0.1693956844 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000049_160 |
0.1699861689 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 20 |
Hb_000181_460 |
0.1706975575 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |