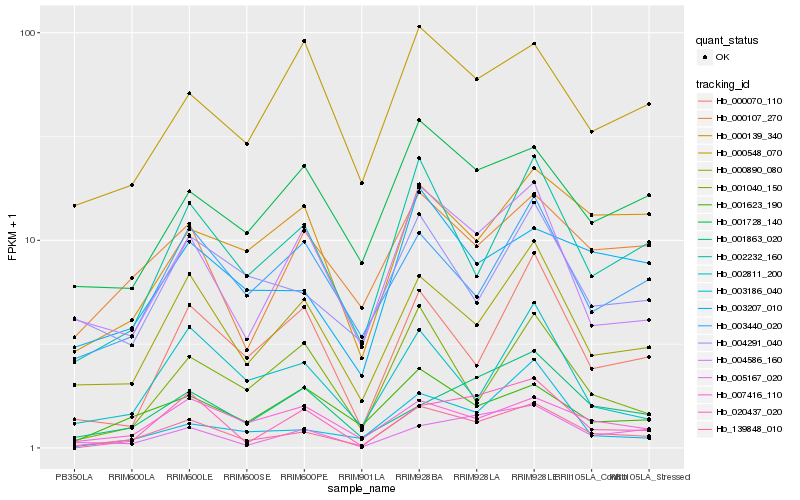
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139848_010 |
0.0 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 2 |
Hb_000890_080 |
0.1347453413 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 3 |
Hb_002232_160 |
0.1437867707 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 4 |
Hb_004586_160 |
0.1453875956 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007416_110 |
0.1689564798 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003207_010 |
0.1694244643 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 7 |
Hb_020437_020 |
0.1722617637 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 8 |
Hb_000107_270 |
0.1767406549 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000070_110 |
0.1772879496 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001623_190 |
0.1797405479 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 11 |
Hb_005167_020 |
0.1802377188 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 12 |
Hb_001728_140 |
0.181717655 |
- |
- |
- |
| 13 |
Hb_002811_200 |
0.1874390188 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 14 |
Hb_001863_020 |
0.1874575035 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001040_150 |
0.1874606443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003186_040 |
0.1879118375 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 17 |
Hb_003440_020 |
0.1881005914 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_000548_070 |
0.1907663644 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 19 |
Hb_004291_040 |
0.1912242327 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_000139_340 |
0.192504194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |