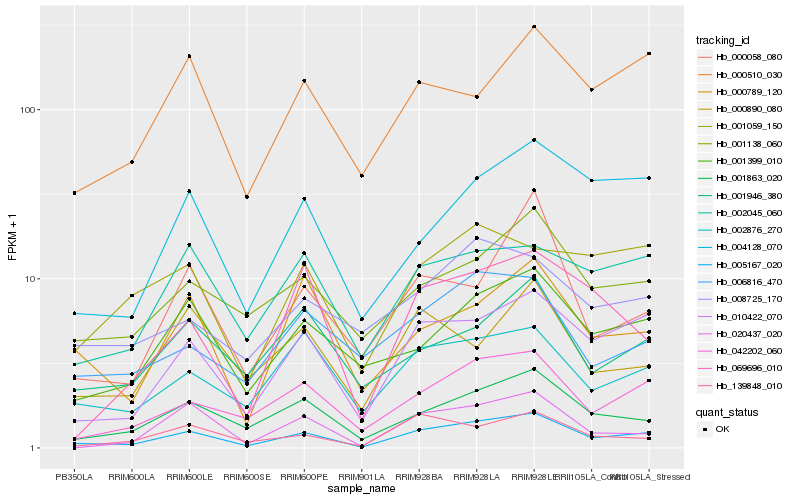
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005167_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 2 |
Hb_010422_070 |
0.1234725026 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 3 |
Hb_001863_020 |
0.1401736899 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_042202_060 |
0.1423552221 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 5 |
Hb_002876_270 |
0.1609306456 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001399_010 |
0.1635375219 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_004128_070 |
0.1659765314 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_020437_020 |
0.1715504836 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 9 |
Hb_001138_060 |
0.1725131112 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 10 |
Hb_000789_120 |
0.1745990695 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002045_060 |
0.1801649502 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_139848_010 |
0.1802377188 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 13 |
Hb_000510_030 |
0.1827811421 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 14 |
Hb_000058_080 |
0.1836167259 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_006816_470 |
0.1838770818 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001946_380 |
0.1852007727 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 17 |
Hb_069696_010 |
0.1862753957 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 18 |
Hb_001059_150 |
0.1904161857 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 19 |
Hb_000890_080 |
0.1939087778 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 20 |
Hb_008725_170 |
0.193933098 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |