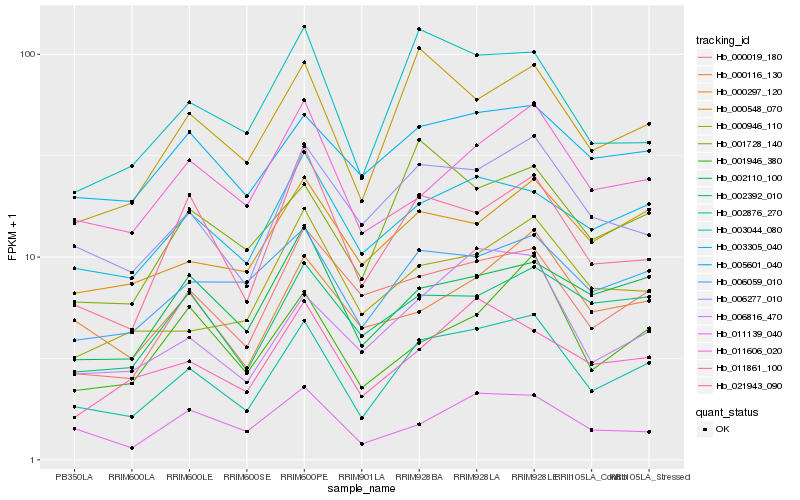
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_270 |
0.0 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000548_070 |
0.1076104878 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_006277_010 |
0.1081044372 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 4 |
Hb_006816_470 |
0.1149622322 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003044_080 |
0.1190018127 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 6 |
Hb_021943_090 |
0.1213729927 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 7 |
Hb_006059_010 |
0.1227859808 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 8 |
Hb_003305_040 |
0.1228398865 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 9 |
Hb_000019_180 |
0.1250057726 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 10 |
Hb_001946_380 |
0.1283006011 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 11 |
Hb_001728_140 |
0.1285667497 |
- |
- |
- |
| 12 |
Hb_011861_100 |
0.1289121386 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000946_110 |
0.1294072589 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 14 |
Hb_000297_120 |
0.1301896493 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 15 |
Hb_002392_010 |
0.1302203531 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 16 |
Hb_005601_040 |
0.1305159025 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 17 |
Hb_011606_020 |
0.1310017613 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 18 |
Hb_011139_040 |
0.1310456886 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002110_100 |
0.131168833 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000116_130 |
0.1319941387 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |