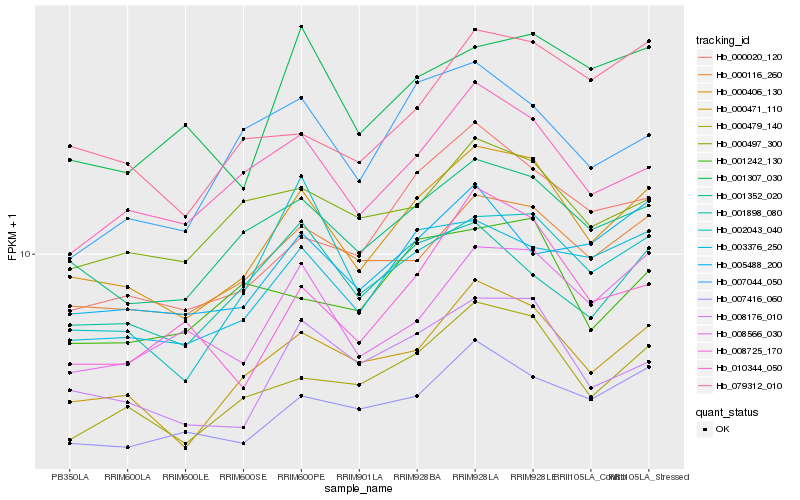
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000406_130 |
0.0 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 2 |
Hb_000471_110 |
0.0619863014 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001352_020 |
0.070317697 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000479_140 |
0.0745951744 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 5 |
Hb_008176_010 |
0.0772968708 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 6 |
Hb_000116_260 |
0.0783104125 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 7 |
Hb_003376_250 |
0.0818832867 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 8 |
Hb_010344_050 |
0.0875768149 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 9 |
Hb_007416_060 |
0.0877564225 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 10 |
Hb_000020_120 |
0.0919616734 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_008566_030 |
0.093825518 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_005488_200 |
0.1002824172 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000497_300 |
0.1004053993 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 14 |
Hb_002043_040 |
0.101606322 |
- |
- |
PREDICTED: uncharacterized protein LOC105631509 [Jatropha curcas] |
| 15 |
Hb_079312_010 |
0.1023588082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001307_030 |
0.1046905725 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 17 |
Hb_008725_170 |
0.1071526905 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 18 |
Hb_001242_130 |
0.1075800465 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 19 |
Hb_007044_050 |
0.1096259602 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 20 |
Hb_001898_080 |
0.1109182668 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |