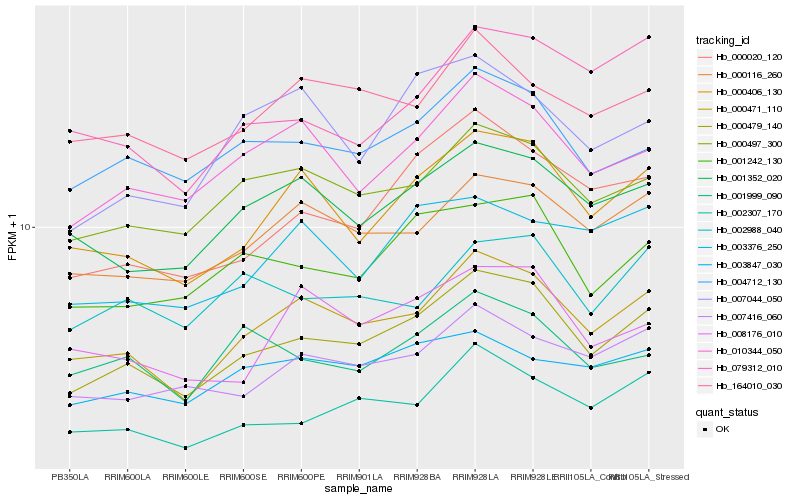
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_140 |
0.0 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 2 |
Hb_000471_110 |
0.0603004071 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000406_130 |
0.0745951744 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 4 |
Hb_000020_120 |
0.0818728255 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001242_130 |
0.085727598 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 6 |
Hb_010344_050 |
0.0904151271 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 7 |
Hb_000497_300 |
0.0907696888 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 8 |
Hb_000116_260 |
0.0910572156 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 9 |
Hb_002307_170 |
0.0912782556 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 10 |
Hb_004712_130 |
0.0912820113 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 11 |
Hb_001352_020 |
0.0943158566 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_007416_060 |
0.0943463907 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 13 |
Hb_079312_010 |
0.1052329866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008176_010 |
0.108331458 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 15 |
Hb_003847_030 |
0.1088757661 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002988_040 |
0.1095956734 |
- |
- |
- |
| 17 |
Hb_164010_030 |
0.1106746141 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 18 |
Hb_003376_250 |
0.1113628306 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 19 |
Hb_001999_090 |
0.1114302864 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007044_050 |
0.1114476432 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |