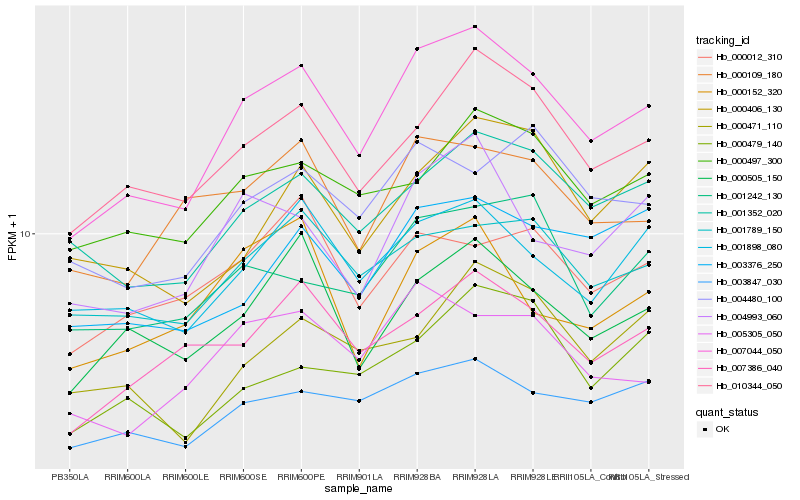
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_050 |
0.0 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 2 |
Hb_010344_050 |
0.0797862553 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 3 |
Hb_004993_060 |
0.0927888805 |
- |
- |
- |
| 4 |
Hb_001352_020 |
0.0931353453 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000505_150 |
0.0933931608 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 6 |
Hb_001898_080 |
0.0934435396 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007386_040 |
0.0947919072 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_001789_150 |
0.0964468623 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003847_030 |
0.0967167925 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Jatropha curcas] |
| 10 |
Hb_005305_050 |
0.0994510616 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 11 |
Hb_000497_300 |
0.1008516811 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 12 |
Hb_000109_180 |
0.1048202458 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 13 |
Hb_001242_130 |
0.1055499967 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 14 |
Hb_003376_250 |
0.1061164481 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 15 |
Hb_004480_100 |
0.107610606 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 16 |
Hb_000012_310 |
0.1081409449 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000406_130 |
0.1096259602 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000479_140 |
0.1114476432 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 19 |
Hb_000471_110 |
0.1124189919 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000152_320 |
0.1124497479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |