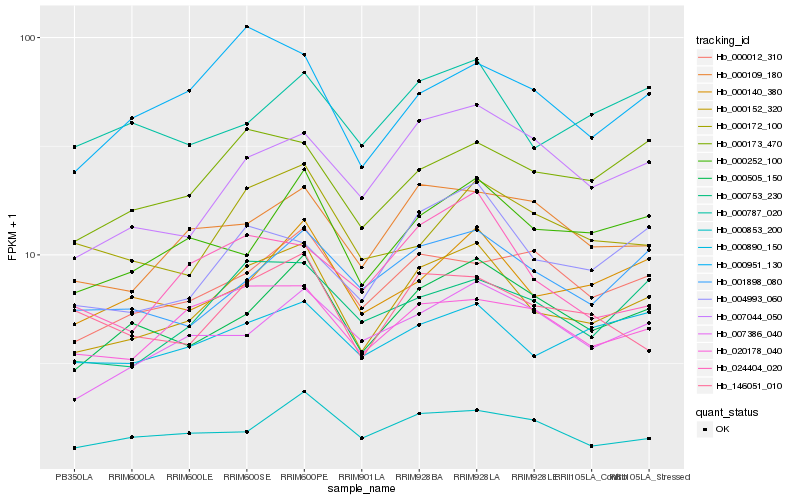
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_320 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000505_150 |
0.0899729871 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 3 |
Hb_000140_380 |
0.1028070321 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 4 |
Hb_004993_060 |
0.1045969838 |
- |
- |
- |
| 5 |
Hb_000753_230 |
0.1061201539 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000252_100 |
0.1064774838 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 7 |
Hb_000173_470 |
0.1076467445 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_020178_040 |
0.1087611249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001898_080 |
0.1098801992 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000109_180 |
0.1103934 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 11 |
Hb_000890_150 |
0.110711107 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 12 |
Hb_007044_050 |
0.1124497479 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 13 |
Hb_007386_040 |
0.1143795314 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000951_130 |
0.1150195453 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 15 |
Hb_000012_310 |
0.1161299725 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_146051_010 |
0.1170035554 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_000787_020 |
0.1173281087 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000853_200 |
0.1175807172 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 19 |
Hb_000172_100 |
0.119002233 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 20 |
Hb_024404_020 |
0.1193762202 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |