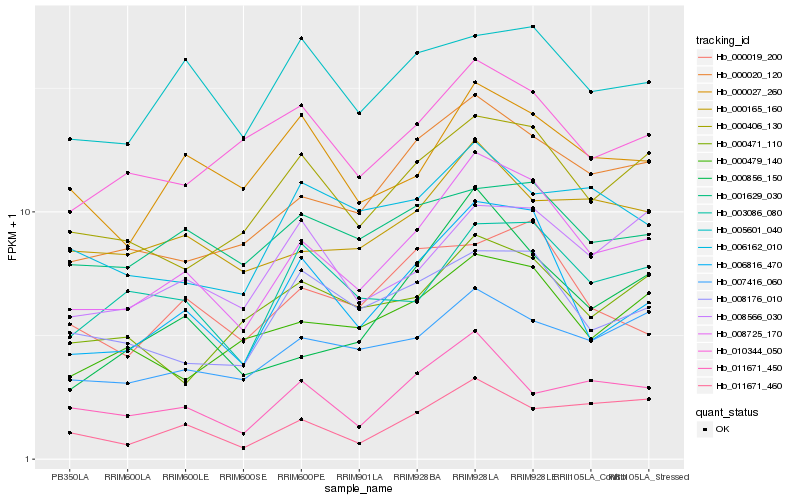
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_170 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 2 |
Hb_006816_470 |
0.08640415 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000020_120 |
0.0928706848 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000406_130 |
0.1071526905 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 5 |
Hb_007416_060 |
0.1118481558 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 6 |
Hb_000479_140 |
0.1185451583 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 7 |
Hb_003086_080 |
0.1192063415 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 8 |
Hb_008176_010 |
0.1198095156 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 9 |
Hb_000165_160 |
0.1222999402 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 10 |
Hb_000856_150 |
0.1238593379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 11 |
Hb_008566_030 |
0.124240549 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_010344_050 |
0.1249058567 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 13 |
Hb_000027_260 |
0.1249285198 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 14 |
Hb_011671_450 |
0.1296447238 |
- |
- |
- |
| 15 |
Hb_011671_460 |
0.1313570325 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 16 |
Hb_000019_200 |
0.1314723898 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_005601_040 |
0.1341750709 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 18 |
Hb_006162_010 |
0.1364199966 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |
| 19 |
Hb_001629_030 |
0.1369930699 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 20 |
Hb_000471_110 |
0.1369967724 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |