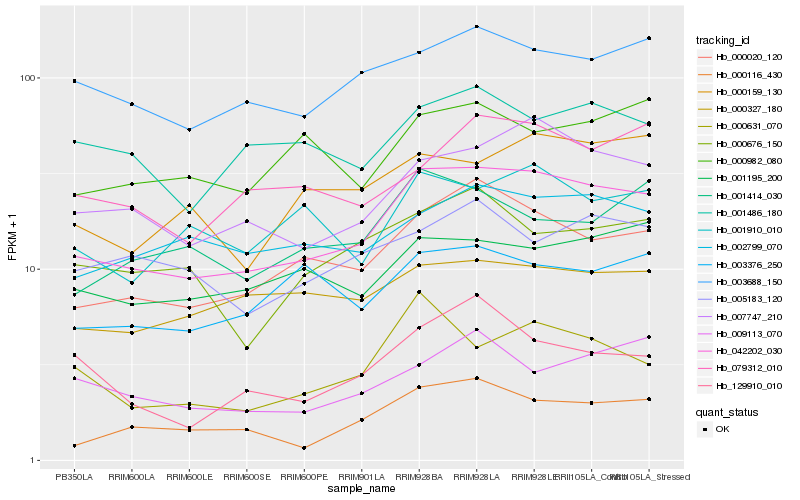
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042202_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 2 |
Hb_007747_210 |
0.0888501586 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000020_120 |
0.0890374704 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_003688_150 |
0.1108427465 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_002799_070 |
0.116736448 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 6 |
Hb_079312_010 |
0.1192742334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000116_430 |
0.1193291867 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001195_200 |
0.1194199442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633802 [Jatropha curcas] |
| 9 |
Hb_000159_130 |
0.1200590066 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000327_180 |
0.1223041404 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001910_010 |
0.1232083085 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_003376_250 |
0.1234422945 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 13 |
Hb_000676_150 |
0.1239285156 |
- |
- |
50S ribosomal protein L7/L12, putative [Ricinus communis] |
| 14 |
Hb_001486_180 |
0.1263088999 |
- |
- |
hypothetical protein B456_007G260200 [Gossypium raimondii] |
| 15 |
Hb_001414_030 |
0.1267463238 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 16 |
Hb_000982_080 |
0.1270910446 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 17 |
Hb_000631_070 |
0.1277906379 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 18 |
Hb_005183_120 |
0.1291682367 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 19 |
Hb_009113_070 |
0.1312548336 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 20 |
Hb_129910_010 |
0.132012981 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1X [Populus euphratica] |