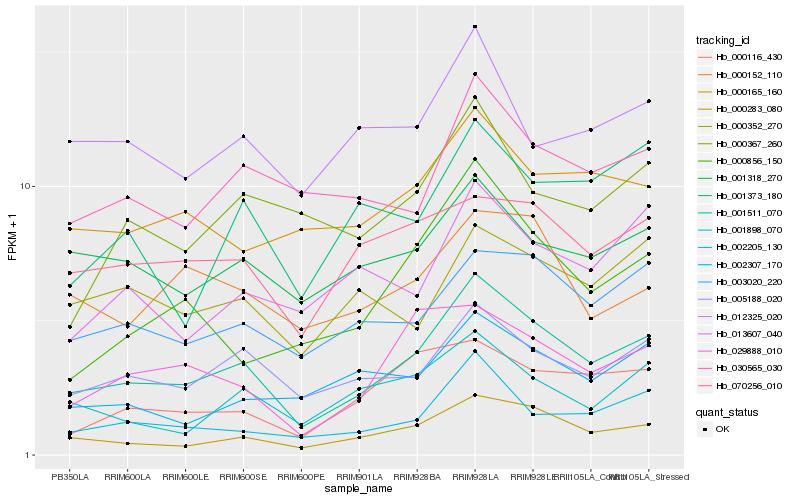
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_070 |
0.0 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 2 |
Hb_000283_080 |
0.0967017995 |
- |
- |
- |
| 3 |
Hb_005188_020 |
0.1208262857 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 4 |
Hb_002205_130 |
0.1233377683 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 5 |
Hb_003020_220 |
0.1304119562 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 6 |
Hb_029888_010 |
0.1361058999 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 7 |
Hb_000856_150 |
0.1389360026 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 8 |
Hb_000116_430 |
0.1420787406 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001898_070 |
0.1430218437 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 10 |
Hb_070256_010 |
0.1443795394 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |
| 11 |
Hb_030565_030 |
0.1451169013 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 12 |
Hb_001318_270 |
0.1455671214 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001373_180 |
0.1473866166 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 14 |
Hb_013607_040 |
0.1493125622 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 15 |
Hb_002307_170 |
0.1493290429 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 16 |
Hb_000165_160 |
0.1498001939 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 17 |
Hb_012325_020 |
0.15217327 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000352_270 |
0.1524613951 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000152_110 |
0.1526700396 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_000367_260 |
0.1536754092 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |