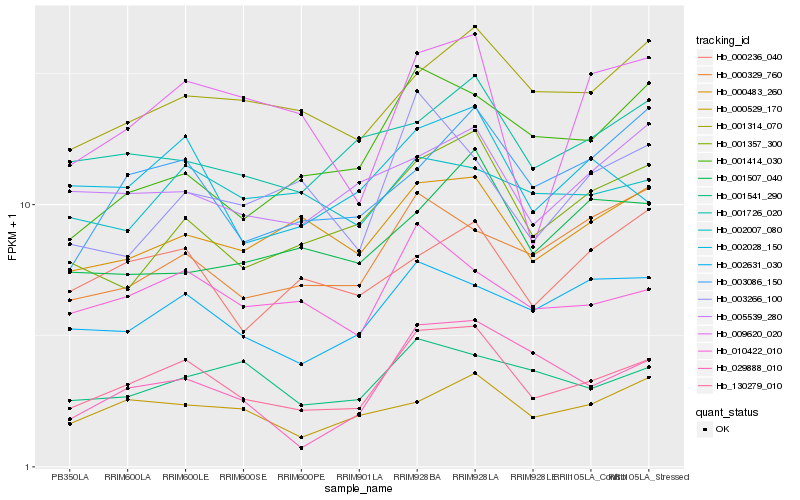
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130279_010 |
0.0 |
- |
- |
PREDICTED: ATP-dependent DNA helicase SRS2-like protein At4g25120 [Jatropha curcas] |
| 2 |
Hb_010422_010 |
0.110967581 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 3 |
Hb_002028_150 |
0.1119594458 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 4 |
Hb_002631_030 |
0.1128412683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001357_300 |
0.1140977066 |
- |
- |
PREDICTED: imidazole glycerol phosphate synthase hisHF, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000483_260 |
0.1167373574 |
- |
- |
PREDICTED: dihydroorotase, mitochondrial isoform X1 [Populus euphratica] |
| 7 |
Hb_001314_070 |
0.1202116164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000529_170 |
0.1239704494 |
- |
- |
hypothetical protein JCGZ_14691 [Jatropha curcas] |
| 9 |
Hb_003086_150 |
0.125979154 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 10 |
Hb_001726_020 |
0.1273435342 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 11 |
Hb_001541_290 |
0.1278176452 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 12 |
Hb_005539_280 |
0.1297465674 |
- |
- |
PREDICTED: uncharacterized protein LOC105644562 [Jatropha curcas] |
| 13 |
Hb_001414_030 |
0.1306579173 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 14 |
Hb_009620_020 |
0.1307380056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000329_760 |
0.1319957555 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 16 |
Hb_000236_040 |
0.1347130112 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 17 |
Hb_029888_010 |
0.1360752904 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 18 |
Hb_002007_080 |
0.1386796108 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 19 |
Hb_003266_100 |
0.1397570277 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 20 |
Hb_001507_040 |
0.1409231821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |