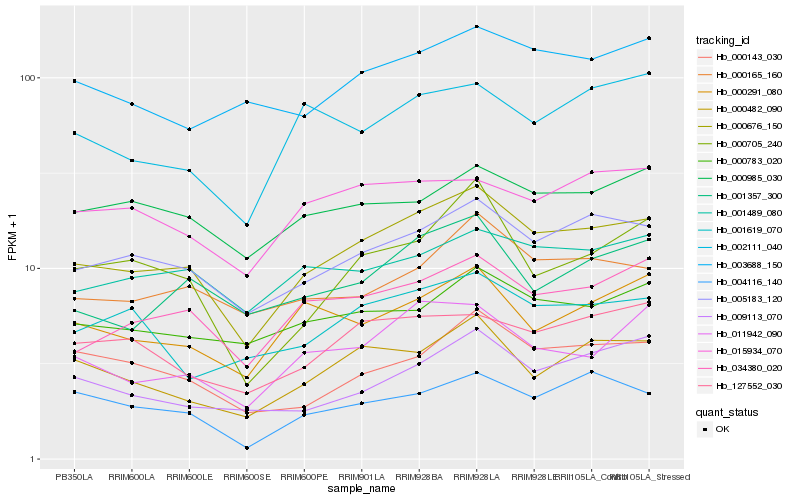
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_150 |
0.0 |
- |
- |
50S ribosomal protein L7/L12, putative [Ricinus communis] |
| 2 |
Hb_005183_120 |
0.071451004 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 3 |
Hb_034380_020 |
0.0909789342 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 4 |
Hb_000482_090 |
0.0958444109 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 5 |
Hb_011942_090 |
0.099878425 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000165_160 |
0.1006907273 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 7 |
Hb_127552_030 |
0.1031542441 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 8 |
Hb_000783_020 |
0.1034267566 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase SDP6, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000143_030 |
0.1040397606 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 10 |
Hb_001357_300 |
0.1041640441 |
- |
- |
PREDICTED: imidazole glycerol phosphate synthase hisHF, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001489_080 |
0.1049284856 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_004116_140 |
0.1052724016 |
- |
- |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 13 |
Hb_000291_080 |
0.1066924764 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 14 |
Hb_000705_240 |
0.1083442408 |
- |
- |
DNA-directed RNA polymerase II 19 kD polypeptide rpb7, putative [Ricinus communis] |
| 15 |
Hb_000985_030 |
0.1091608807 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 16 |
Hb_001619_070 |
0.1093386281 |
- |
- |
PREDICTED: uncharacterized protein LOC103503585 [Cucumis melo] |
| 17 |
Hb_003688_150 |
0.109583963 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 18 |
Hb_015934_070 |
0.1103843709 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009113_070 |
0.1123525837 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 20 |
Hb_002111_040 |
0.1123643213 |
- |
- |
hypothetical protein JCGZ_16989 [Jatropha curcas] |