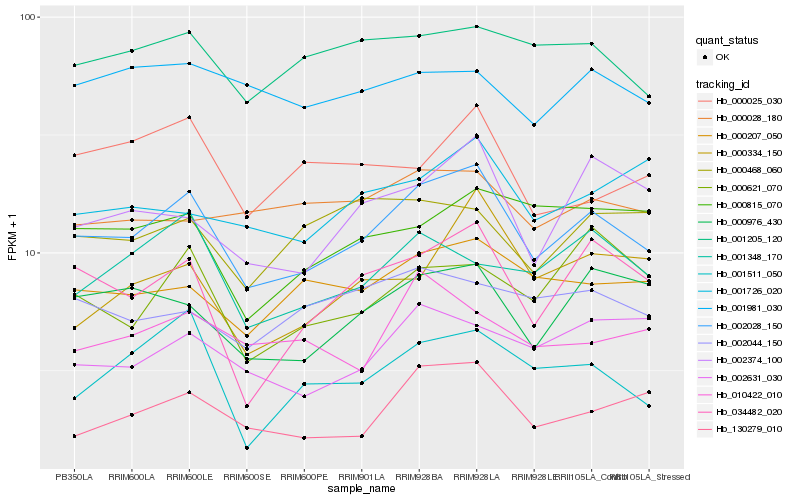
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_150 |
0.0 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 2 |
Hb_034482_020 |
0.1033658544 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000976_430 |
0.1083854312 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJD5 [Jatropha curcas] |
| 4 |
Hb_000207_050 |
0.1088627448 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 5 |
Hb_130279_010 |
0.1119594458 |
- |
- |
PREDICTED: ATP-dependent DNA helicase SRS2-like protein At4g25120 [Jatropha curcas] |
| 6 |
Hb_002374_100 |
0.1138432935 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 7 |
Hb_001205_120 |
0.1149208454 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 8 |
Hb_001348_170 |
0.1194950139 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_002631_030 |
0.1233779952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002044_150 |
0.1241414556 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 11 |
Hb_000815_070 |
0.1241586289 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 12 |
Hb_001511_050 |
0.126138302 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase bub1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000621_070 |
0.1262653986 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 14 |
Hb_000025_030 |
0.1263837683 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001726_020 |
0.1268713177 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 16 |
Hb_000028_180 |
0.1274553992 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010422_010 |
0.1275846465 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_001981_030 |
0.1282307552 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000334_150 |
0.1285424225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000468_060 |
0.1293869567 |
- |
- |
ku P70 DNA helicase, putative [Ricinus communis] |