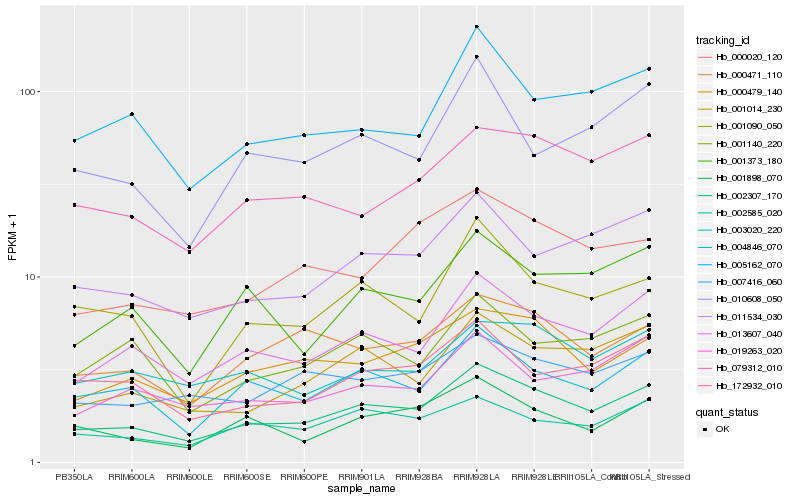
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 2 |
Hb_013607_040 |
0.0717985251 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 3 |
Hb_004846_070 |
0.0887867876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 4 |
Hb_011534_030 |
0.0905168458 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 5 |
Hb_000479_140 |
0.0912782556 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 6 |
Hb_001014_230 |
0.0921255922 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 7 |
Hb_000471_110 |
0.0995219826 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002585_020 |
0.1016134226 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |
| 9 |
Hb_079312_010 |
0.1033357287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001090_050 |
0.103598523 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005162_070 |
0.1041969001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001373_180 |
0.1048221632 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007416_060 |
0.1051719713 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 14 |
Hb_172932_010 |
0.106145558 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000020_120 |
0.1068993028 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001898_070 |
0.1073056568 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 17 |
Hb_003020_220 |
0.1084662768 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010608_050 |
0.110306521 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 19 |
Hb_001140_220 |
0.110550528 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_019263_020 |
0.1115509264 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |