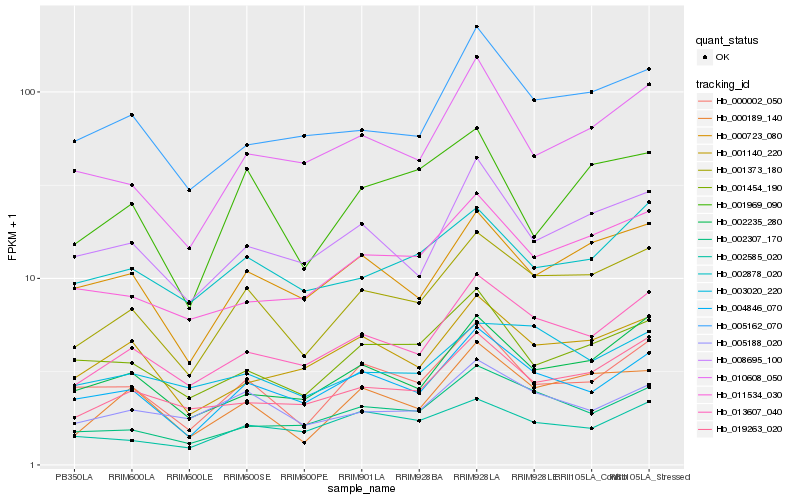
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001373_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 2 |
Hb_013607_040 |
0.0770938849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 3 |
Hb_000189_140 |
0.0860804887 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_004846_070 |
0.0930711992 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 5 |
Hb_000723_080 |
0.094140566 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 6 |
Hb_000002_050 |
0.0976218133 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 7 |
Hb_002235_280 |
0.1024234197 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15300 [Jatropha curcas] |
| 8 |
Hb_019263_020 |
0.1026017589 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002307_170 |
0.1048221632 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 10 |
Hb_008695_100 |
0.1065553654 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_011534_030 |
0.1065630474 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 12 |
Hb_010608_050 |
0.1127463949 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 13 |
Hb_005162_070 |
0.1132508689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005188_020 |
0.1138645384 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 15 |
Hb_001140_220 |
0.1149142886 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003020_220 |
0.1153664508 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002878_020 |
0.1159009242 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 18 |
Hb_001454_190 |
0.1160597872 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 19 |
Hb_001969_090 |
0.1181422194 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 20 |
Hb_002585_020 |
0.1196818536 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |