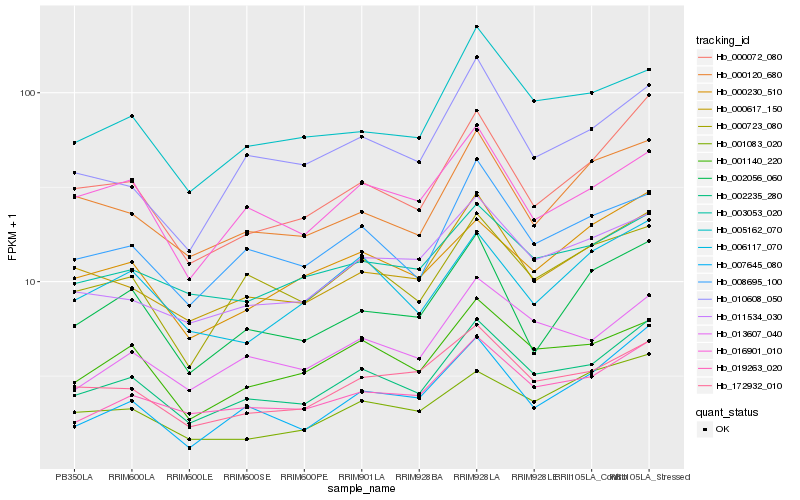
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_280 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15300 [Jatropha curcas] |
| 2 |
Hb_019263_020 |
0.066344536 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000072_080 |
0.0673915168 |
- |
- |
PREDICTED: probable ribosomal protein S11, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000617_150 |
0.0760256696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 5 |
Hb_008695_100 |
0.0769075411 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001140_220 |
0.0769272826 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_013607_040 |
0.0783449114 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 8 |
Hb_172932_010 |
0.0818468384 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000723_080 |
0.0823873013 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 10 |
Hb_011534_030 |
0.0841704617 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 11 |
Hb_005162_070 |
0.087006581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003053_020 |
0.0880213698 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 13 |
Hb_000230_510 |
0.0888654773 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 14 |
Hb_010608_050 |
0.0900098437 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 15 |
Hb_000120_680 |
0.0915868547 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 16 |
Hb_006117_070 |
0.0947441285 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 17 |
Hb_016901_010 |
0.0962633921 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 18 |
Hb_007645_080 |
0.0966252027 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 19 |
Hb_002056_060 |
0.0972518203 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001083_020 |
0.0977781438 |
- |
- |
- |