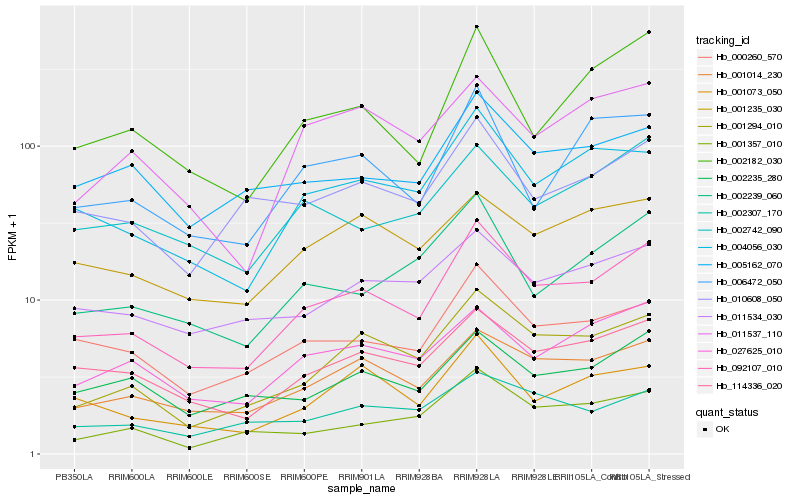
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092107_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001014_230 |
0.0921535813 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 3 |
Hb_001294_010 |
0.0947875115 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 4 |
Hb_004056_030 |
0.0968170355 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 5 |
Hb_114336_020 |
0.0995364312 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 6 |
Hb_000260_570 |
0.1003125666 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002182_030 |
0.1130552637 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 8 |
Hb_027625_010 |
0.1145062118 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 9 |
Hb_002239_060 |
0.1154625146 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 10 |
Hb_001073_050 |
0.1186880087 |
- |
- |
- |
| 11 |
Hb_001357_010 |
0.1198444469 |
- |
- |
- |
| 12 |
Hb_010608_050 |
0.1199296012 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 13 |
Hb_011534_030 |
0.1220618738 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 14 |
Hb_006472_050 |
0.1224298445 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_002742_090 |
0.1238794134 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 16 |
Hb_002235_280 |
0.1244207157 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15300 [Jatropha curcas] |
| 17 |
Hb_001235_030 |
0.1249928995 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 18 |
Hb_011537_110 |
0.1257939927 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 19 |
Hb_002307_170 |
0.1261552446 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_005162_070 |
0.1271683689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |