| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
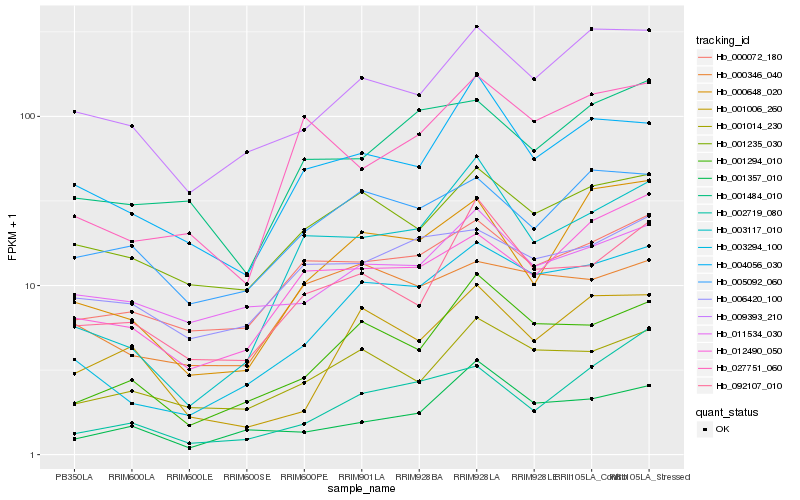
Hb_003294_100 |
0.0 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 2 |
Hb_001294_010 |
0.1162632401 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 3 |
Hb_003117_010 |
0.1328339588 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 4 |
Hb_009393_210 |
0.1442480735 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 5 |
Hb_000648_020 |
0.1450550703 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 6 |
Hb_012490_050 |
0.155279333 |
- |
- |
PREDICTED: uncharacterized protein LOC105637921 [Jatropha curcas] |
| 7 |
Hb_001014_230 |
0.1565216717 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 8 |
Hb_027751_060 |
0.1602759118 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 9 |
Hb_092107_010 |
0.161070357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000072_180 |
0.1635722108 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 11 |
Hb_001357_010 |
0.1642387087 |
- |
- |
- |
| 12 |
Hb_004056_030 |
0.1655106467 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 13 |
Hb_001484_010 |
0.1657778899 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 14 |
Hb_001235_030 |
0.1662957856 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 15 |
Hb_000346_040 |
0.1682004006 |
- |
- |
- |
| 16 |
Hb_005092_060 |
0.1714552693 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 17 |
Hb_002719_080 |
0.1723401509 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 18 |
Hb_011534_030 |
0.1739426753 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 19 |
Hb_001006_260 |
0.1746710682 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 20 |
Hb_006420_100 |
0.1747332246 |
- |
- |
PREDICTED: protein bicaudal C homolog 1 [Jatropha curcas] |