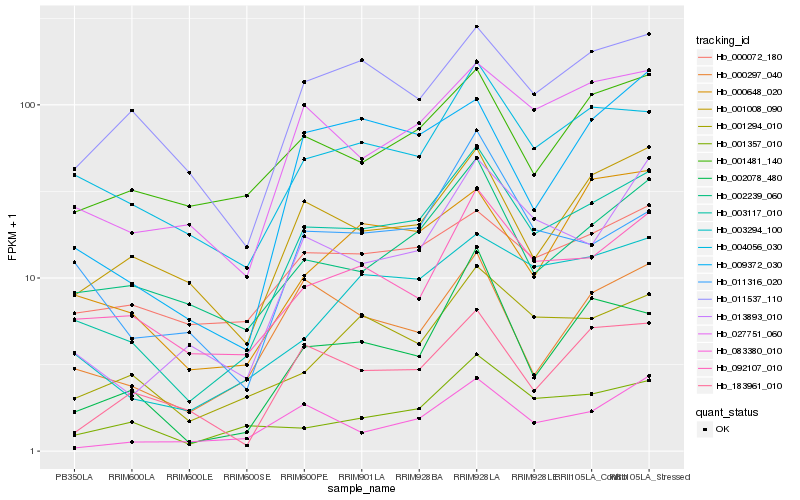
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003117_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 2 |
Hb_027751_060 |
0.1245871714 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 3 |
Hb_003294_100 |
0.1328339588 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 4 |
Hb_013893_010 |
0.1350636446 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 5 |
Hb_092107_010 |
0.13903701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001294_010 |
0.1409225962 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 7 |
Hb_002078_480 |
0.1458052759 |
- |
- |
hypothetical protein CICLE_v10033318mg [Citrus clementina] |
| 8 |
Hb_004056_030 |
0.146849769 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 9 |
Hb_009372_030 |
0.1510728302 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 10 |
Hb_083380_010 |
0.1533658595 |
- |
- |
- |
| 11 |
Hb_000297_040 |
0.1539272713 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 12 |
Hb_002239_060 |
0.1550781648 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 13 |
Hb_001008_090 |
0.1574425457 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 14 |
Hb_011316_020 |
0.159171499 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 15 |
Hb_000648_020 |
0.1621505044 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 16 |
Hb_001481_140 |
0.1634529683 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 17 |
Hb_011537_110 |
0.1643369803 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 18 |
Hb_001357_010 |
0.1643603655 |
- |
- |
- |
| 19 |
Hb_183961_010 |
0.1644843805 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 20 |
Hb_000072_180 |
0.1652802403 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |