| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
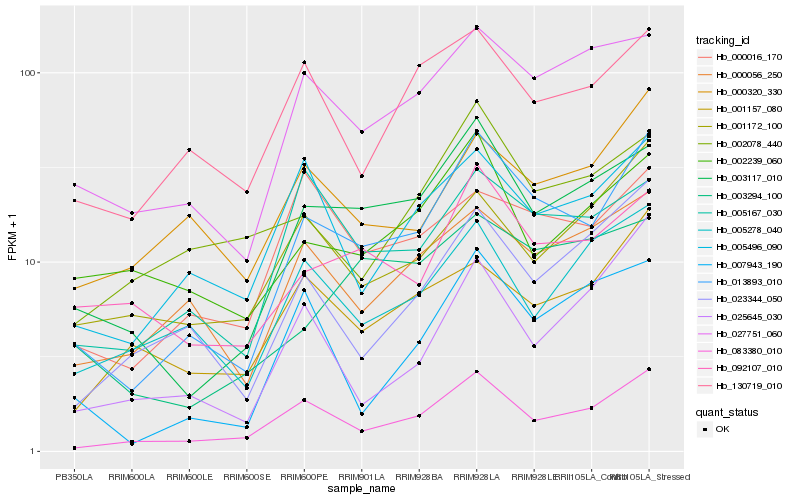
Hb_013893_010 |
0.0 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 2 |
Hb_083380_010 |
0.1320361496 |
- |
- |
- |
| 3 |
Hb_003117_010 |
0.1350636446 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 4 |
Hb_027751_060 |
0.1564778292 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 5 |
Hb_000056_250 |
0.1669740276 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 6 |
Hb_092107_010 |
0.1678802691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_023344_050 |
0.1707185694 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 8 |
Hb_005496_090 |
0.1750932504 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_005167_030 |
0.17548598 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 10 |
Hb_007943_190 |
0.17593302 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 11 |
Hb_130719_010 |
0.1798714744 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 12 |
Hb_000320_330 |
0.182327075 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 13 |
Hb_000016_170 |
0.1824471341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001172_100 |
0.1859085464 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001157_080 |
0.1863504633 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_003294_100 |
0.1868353167 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 17 |
Hb_002078_440 |
0.1885067896 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005278_040 |
0.1895780205 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 19 |
Hb_002239_060 |
0.1915959475 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 20 |
Hb_025645_030 |
0.1935764027 |
- |
- |
- |