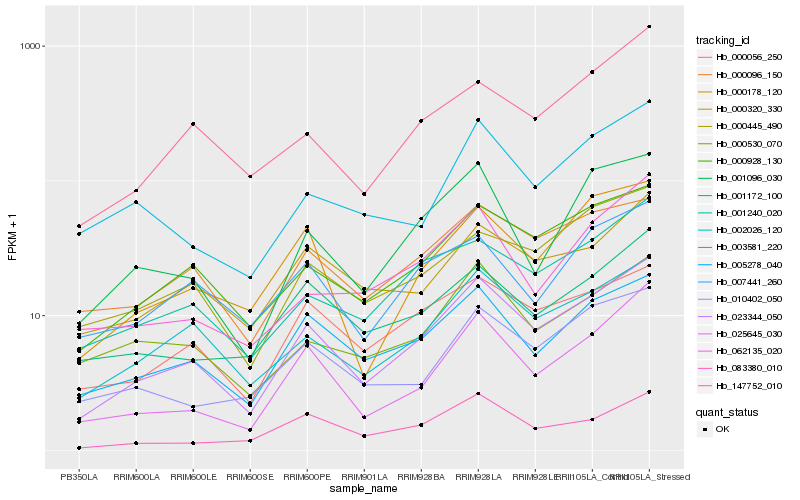
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023344_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 2 |
Hb_000928_130 |
0.0974010147 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_002026_120 |
0.0980517582 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 4 |
Hb_025645_030 |
0.1020683035 |
- |
- |
- |
| 5 |
Hb_005278_040 |
0.1051098544 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 6 |
Hb_001240_020 |
0.111388417 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 7 |
Hb_000056_250 |
0.1167644257 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_001096_030 |
0.1197760278 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 9 |
Hb_000178_120 |
0.1203622479 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 10 |
Hb_000445_490 |
0.1210173399 |
- |
- |
- |
| 11 |
Hb_000096_150 |
0.1221187898 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_147752_010 |
0.1240817597 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 13 |
Hb_001172_100 |
0.1251873445 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_010402_050 |
0.1252543738 |
- |
- |
- |
| 15 |
Hb_000320_330 |
0.1288646283 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 16 |
Hb_083380_010 |
0.1301637641 |
- |
- |
- |
| 17 |
Hb_000530_070 |
0.1309604791 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 18 |
Hb_062135_020 |
0.1321463771 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 19 |
Hb_007441_260 |
0.1336234892 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_003581_220 |
0.1345451998 |
- |
- |
unnamed protein product [Vitis vinifera] |