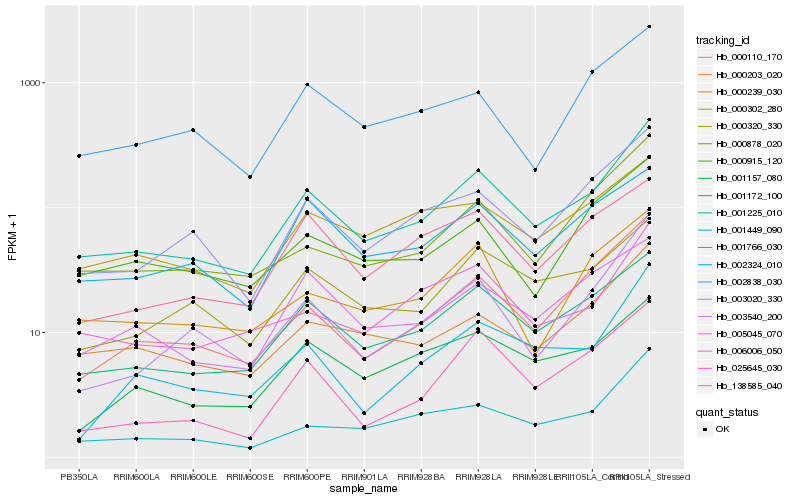
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001225_010 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_001172_100 |
0.0820979567 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003020_330 |
0.0996438601 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 4 |
Hb_000110_170 |
0.1018664794 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000203_020 |
0.1028279842 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 6 |
Hb_001449_090 |
0.112507056 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000878_020 |
0.114011386 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 8 |
Hb_006006_050 |
0.1151311972 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 9 |
Hb_003540_200 |
0.1161184611 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 10 |
Hb_000915_120 |
0.1190407099 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 11 |
Hb_000320_330 |
0.1192811971 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 12 |
Hb_002324_010 |
0.1202227577 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 13 |
Hb_001157_080 |
0.1225100534 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000239_030 |
0.1226316461 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 15 |
Hb_005045_070 |
0.123207895 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 16 |
Hb_002838_030 |
0.1245975942 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 17 |
Hb_000302_280 |
0.1254822977 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 18 |
Hb_138585_040 |
0.1261316588 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 19 |
Hb_025645_030 |
0.1275114421 |
- |
- |
- |
| 20 |
Hb_001766_030 |
0.1285043594 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |