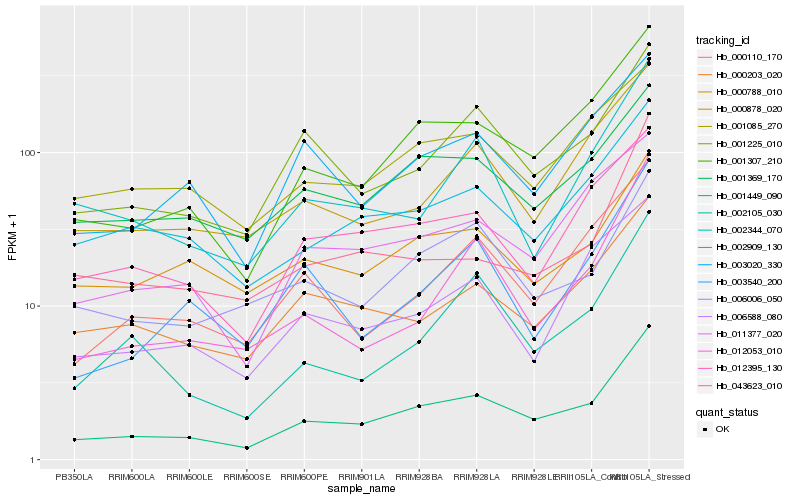
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001449_090 |
0.0 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001307_210 |
0.0773504923 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 3 |
Hb_001225_010 |
0.112507056 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_002909_130 |
0.1177413512 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_012395_130 |
0.1210597065 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 6 |
Hb_011377_020 |
0.1217601466 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 7 |
Hb_000878_020 |
0.1220392398 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 8 |
Hb_000110_170 |
0.1250477557 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_003020_330 |
0.125713186 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 10 |
Hb_001369_170 |
0.1258865975 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 11 |
Hb_000203_020 |
0.1270090855 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_002105_030 |
0.1288883899 |
- |
- |
- |
| 13 |
Hb_006006_050 |
0.1308890788 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 14 |
Hb_002344_070 |
0.1349022955 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 15 |
Hb_006588_080 |
0.1358442988 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 16 |
Hb_012053_010 |
0.1367696576 |
- |
- |
srpk, putative [Ricinus communis] |
| 17 |
Hb_000788_010 |
0.1375497463 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 18 |
Hb_043623_010 |
0.1375504226 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 19 |
Hb_001085_270 |
0.1381900616 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 20 |
Hb_003540_200 |
0.1384619261 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |