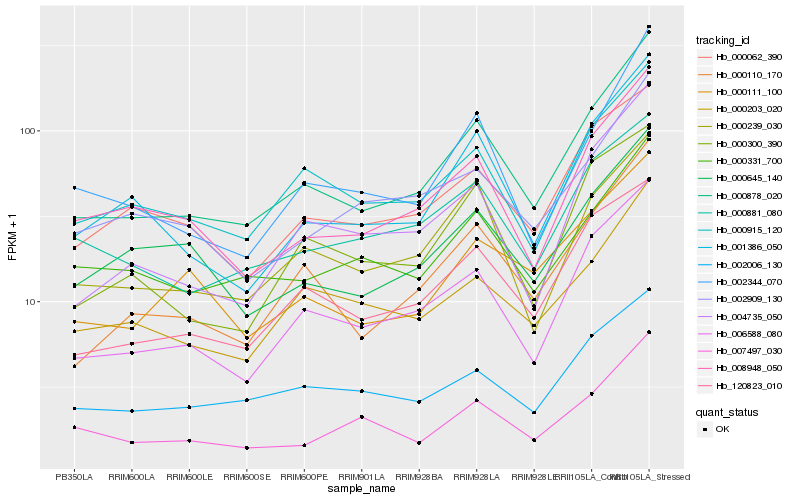
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 2 |
Hb_006588_080 |
0.0709779881 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 3 |
Hb_000110_170 |
0.0734083775 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_001386_050 |
0.0803863796 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_008948_050 |
0.0882412289 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 6 |
Hb_002344_070 |
0.0892228361 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 7 |
Hb_000915_120 |
0.0918665624 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 8 |
Hb_004735_050 |
0.0925571561 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 9 |
Hb_120823_010 |
0.0936960115 |
- |
- |
- |
| 10 |
Hb_000111_100 |
0.1035935132 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 11 |
Hb_000203_020 |
0.103624265 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_000239_030 |
0.1054833652 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 13 |
Hb_000062_390 |
0.1085318321 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002006_130 |
0.10860524 |
- |
- |
PREDICTED: uncharacterized protein LOC105648712 [Jatropha curcas] |
| 15 |
Hb_000331_700 |
0.1087721851 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 16 |
Hb_002909_130 |
0.1089420881 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_007497_030 |
0.1095225389 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 18 |
Hb_000881_080 |
0.1098920625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000645_140 |
0.1104033508 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A [Nelumbo nucifera] |
| 20 |
Hb_000300_390 |
0.11254741 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |