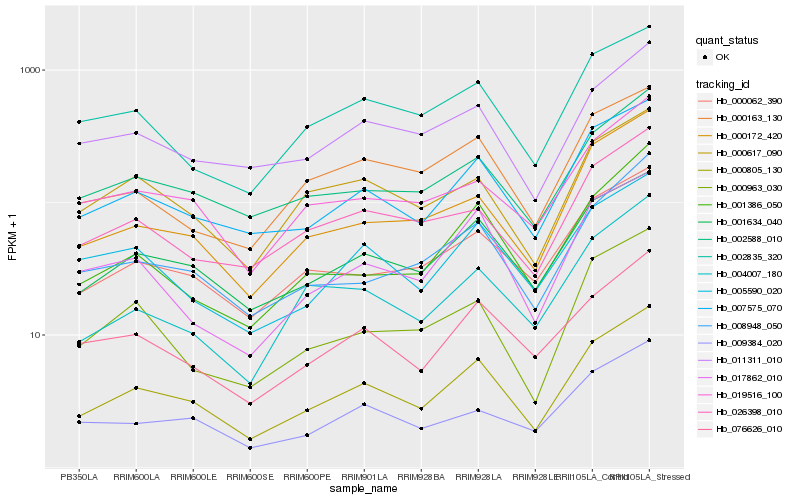
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007575_070 |
0.0667534646 |
- |
- |
Histone H3.2 [Glycine soja] |
| 3 |
Hb_001634_040 |
0.0711192933 |
transcription factor |
TF Family: NF-YC |
DNA polymerase epsilon subunit, putative [Ricinus communis] |
| 4 |
Hb_000172_420 |
0.0884139032 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 5 |
Hb_000062_390 |
0.0930725411 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 6 |
Hb_008948_050 |
0.0935165875 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 7 |
Hb_076626_010 |
0.0943648471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002588_010 |
0.0977013868 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 9 |
Hb_017862_010 |
0.0984179107 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 10 |
Hb_001386_050 |
0.0989764221 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 11 |
Hb_019516_100 |
0.0999091722 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 12 |
Hb_000963_030 |
0.1000711546 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 13 |
Hb_004007_180 |
0.1001342219 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 14 |
Hb_005590_020 |
0.1008231033 |
- |
- |
hypothetical protein CICLE_v10010008mg [Citrus clementina] |
| 15 |
Hb_002835_320 |
0.101247138 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 16 |
Hb_026398_010 |
0.1018902738 |
- |
- |
hypothetical protein B456_010G132100 [Gossypium raimondii] |
| 17 |
Hb_000163_130 |
0.102721803 |
- |
- |
hypothetical protein CICLE_v10022870mg [Citrus clementina] |
| 18 |
Hb_009384_020 |
0.1032387044 |
- |
- |
hypothetical protein JCGZ_24168 [Jatropha curcas] |
| 19 |
Hb_000617_090 |
0.1039882588 |
- |
- |
hypoia-responsive family protein 4 [Hevea brasiliensis] |
| 20 |
Hb_011311_010 |
0.1042280568 |
- |
- |
60S ribosomal protein L27C [Hevea brasiliensis] |