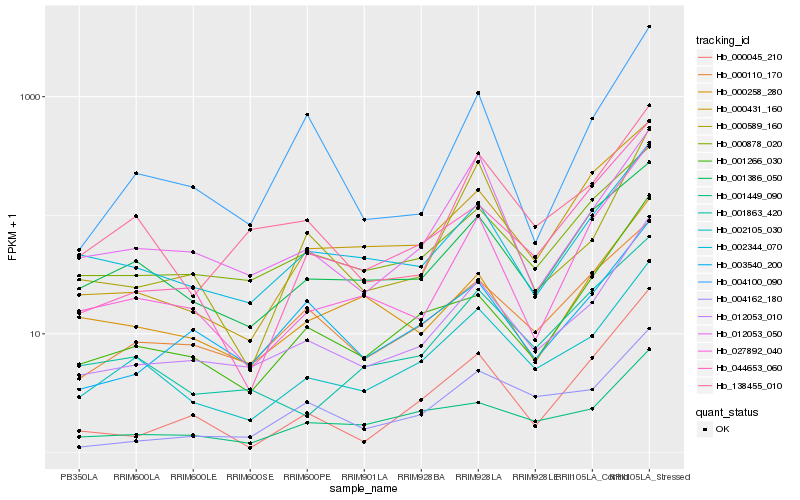
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012053_010 |
0.0 |
- |
- |
srpk, putative [Ricinus communis] |
| 2 |
Hb_002105_030 |
0.1146210847 |
- |
- |
- |
| 3 |
Hb_002344_070 |
0.1170461147 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 4 |
Hb_000878_020 |
0.1183752779 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 5 |
Hb_003540_200 |
0.1188100469 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_001266_030 |
0.120783975 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000110_170 |
0.122140928 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_138455_010 |
0.1253187187 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_027892_040 |
0.1308169991 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 10 |
Hb_044653_060 |
0.1309706381 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 11 |
Hb_001449_090 |
0.1367696576 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000431_160 |
0.139830095 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 13 |
Hb_000258_280 |
0.1406511266 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 14 |
Hb_001386_050 |
0.143541146 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 15 |
Hb_000045_210 |
0.1439619911 |
- |
- |
- |
| 16 |
Hb_012053_050 |
0.1473220369 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 17 |
Hb_004162_180 |
0.1489707276 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_001863_420 |
0.15064818 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 19 |
Hb_004100_090 |
0.1529771658 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 20 |
Hb_000589_160 |
0.154273664 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |