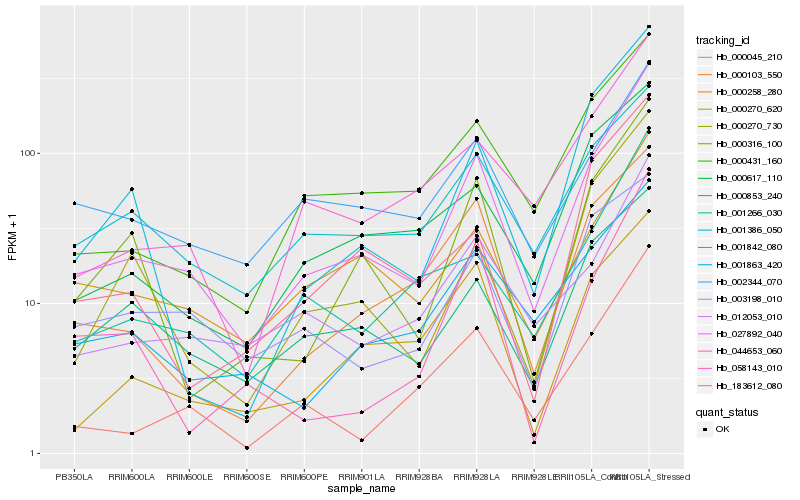
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027892_040 |
0.0 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 2 |
Hb_000045_210 |
0.1286470498 |
- |
- |
- |
| 3 |
Hb_012053_010 |
0.1308169991 |
- |
- |
srpk, putative [Ricinus communis] |
| 4 |
Hb_001266_030 |
0.1345136682 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_044653_060 |
0.1352723546 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 6 |
Hb_000431_160 |
0.1357741831 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 7 |
Hb_000316_100 |
0.1362315771 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000258_280 |
0.146158687 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 9 |
Hb_000270_730 |
0.1475562443 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 10 |
Hb_000617_110 |
0.1514087244 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 11 |
Hb_001842_080 |
0.1519392032 |
- |
- |
- |
| 12 |
Hb_001863_420 |
0.1528042688 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 13 |
Hb_000103_550 |
0.1546595819 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000853_240 |
0.1574771519 |
- |
- |
- |
| 15 |
Hb_000270_620 |
0.1580440294 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_183612_080 |
0.1594706822 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 17 |
Hb_002344_070 |
0.1599860263 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 18 |
Hb_058143_010 |
0.1640717015 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 19 |
Hb_001386_050 |
0.166338965 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 20 |
Hb_003198_010 |
0.1670482164 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |