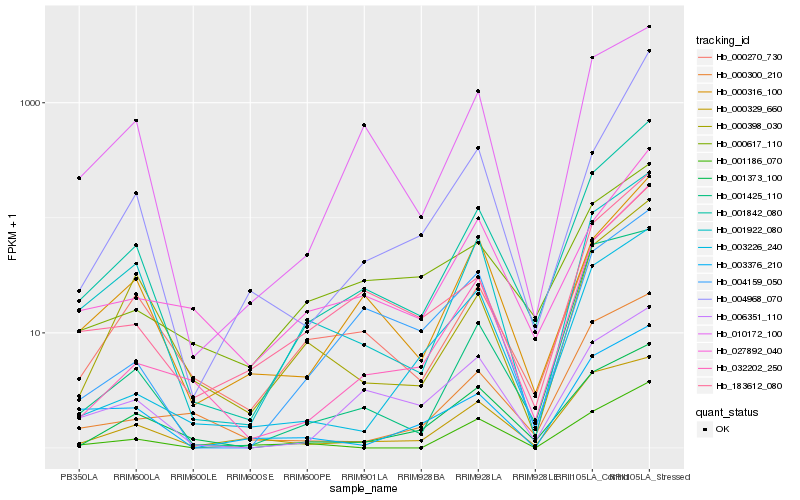
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001842_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_000270_730 |
0.0965544854 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 3 |
Hb_001922_080 |
0.1245951532 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 4 |
Hb_032202_250 |
0.1316895879 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 5 |
Hb_000316_100 |
0.1326632138 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_183612_080 |
0.1418701177 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 7 |
Hb_010172_100 |
0.1484436482 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 8 |
Hb_027892_040 |
0.1519392032 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 9 |
Hb_001373_100 |
0.1551082041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000398_030 |
0.1585123779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000329_660 |
0.1619485885 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006351_110 |
0.1639832804 |
- |
- |
- |
| 13 |
Hb_000300_210 |
0.1643007932 |
- |
- |
gd2b, putative [Ricinus communis] |
| 14 |
Hb_003226_240 |
0.1680514094 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 15 |
Hb_004968_070 |
0.1690528121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004159_050 |
0.1697347257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003376_210 |
0.1733211995 |
- |
- |
- |
| 18 |
Hb_000617_110 |
0.1750652872 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 19 |
Hb_001186_070 |
0.1758555057 |
- |
- |
PREDICTED: F-box protein FBW2-like [Gossypium raimondii] |
| 20 |
Hb_001425_110 |
0.176392751 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |