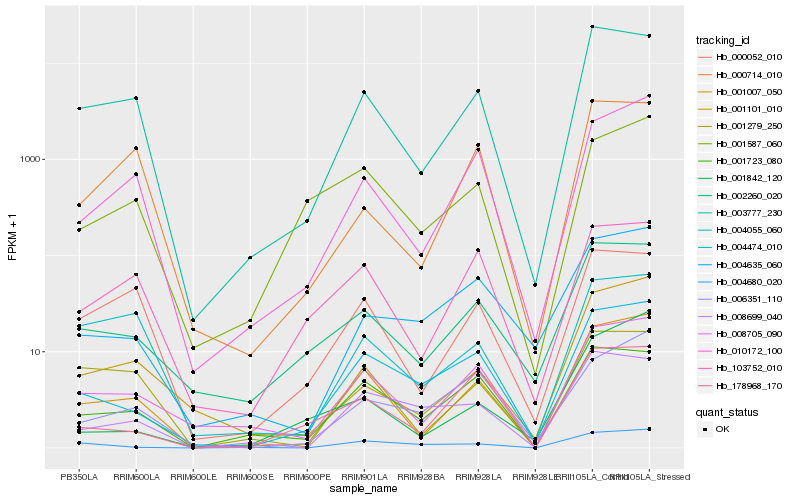
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001007_050 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 2 |
Hb_010172_100 |
0.1281565749 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 3 |
Hb_008705_090 |
0.1285237495 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 4 |
Hb_003777_230 |
0.1318337476 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 5 |
Hb_004474_010 |
0.1473917891 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_001101_010 |
0.1670886574 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 7 |
Hb_002260_020 |
0.1673444527 |
- |
- |
- |
| 8 |
Hb_000052_010 |
0.1825723967 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 9 |
Hb_001723_080 |
0.1840572739 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 10 |
Hb_006351_110 |
0.1868755614 |
- |
- |
- |
| 11 |
Hb_004635_060 |
0.1874752471 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 12 |
Hb_001587_060 |
0.1876316593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001842_120 |
0.1905345287 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_008699_040 |
0.1912824732 |
- |
- |
- |
| 15 |
Hb_001279_250 |
0.1920692638 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 16 |
Hb_178968_170 |
0.1950985407 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 homolog [Jatropha curcas] |
| 17 |
Hb_004055_060 |
0.1956785201 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 18 |
Hb_103752_010 |
0.1964882012 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 19 |
Hb_000714_010 |
0.1996391824 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 20 |
Hb_004680_020 |
0.1996985853 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |