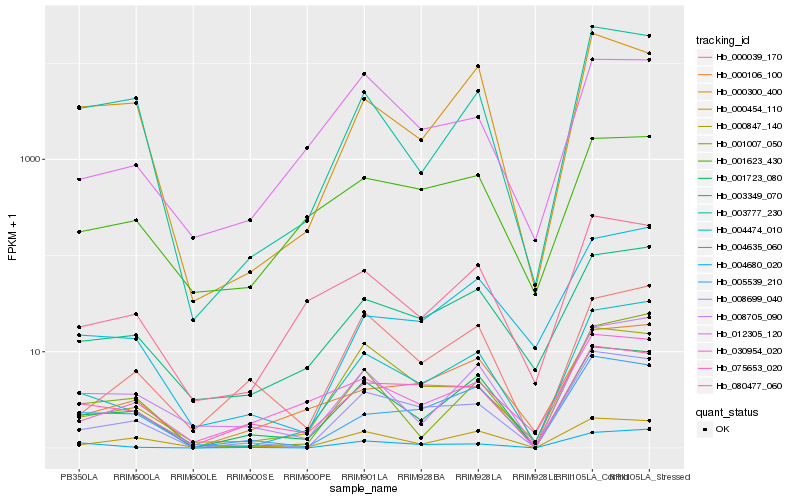
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008699_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_004474_010 |
0.1418279772 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_000847_140 |
0.1421337371 |
- |
- |
- |
| 4 |
Hb_001723_080 |
0.1467091423 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 5 |
Hb_075653_020 |
0.1553562694 |
- |
- |
- |
| 6 |
Hb_005539_210 |
0.1608960601 |
- |
- |
- |
| 7 |
Hb_004680_020 |
0.1644879508 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 8 |
Hb_003777_230 |
0.1726810452 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 9 |
Hb_003349_070 |
0.172920207 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 10 |
Hb_000454_110 |
0.1763085598 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 11 |
Hb_000039_170 |
0.1815996325 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 12 |
Hb_080477_060 |
0.1881086711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008705_090 |
0.1884886578 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 14 |
Hb_012305_120 |
0.1912637875 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 15 |
Hb_001007_050 |
0.1912824732 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 16 |
Hb_001623_430 |
0.1916304756 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 17 |
Hb_030954_020 |
0.1920581022 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 18 |
Hb_004635_060 |
0.1925556349 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 19 |
Hb_000106_100 |
0.1927368328 |
- |
- |
- |
| 20 |
Hb_000300_400 |
0.1979416079 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |