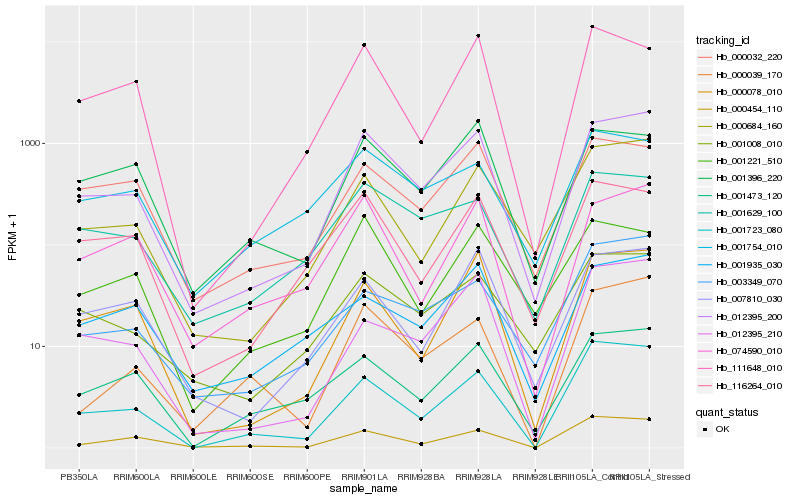
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000684_160 |
0.1219586002 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001008_010 |
0.1262688154 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_000078_010 |
0.1303951627 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 5 |
Hb_001723_080 |
0.1344269936 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 6 |
Hb_007810_030 |
0.1355510797 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 7 |
Hb_074590_010 |
0.1370549744 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 8 |
Hb_000454_110 |
0.1429980492 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 9 |
Hb_001629_100 |
0.1430685127 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X2 [Vitis vinifera] |
| 10 |
Hb_001473_120 |
0.1431887941 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_116264_010 |
0.1437767766 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 12 |
Hb_012395_210 |
0.147881564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000032_220 |
0.1492971904 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001396_220 |
0.1506015691 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 15 |
Hb_003349_070 |
0.1523126924 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 16 |
Hb_000039_170 |
0.1541272418 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 17 |
Hb_001935_030 |
0.1550474026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_111648_010 |
0.1575878291 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 19 |
Hb_001221_510 |
0.1620295738 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 20 |
Hb_001754_010 |
0.1620505059 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |