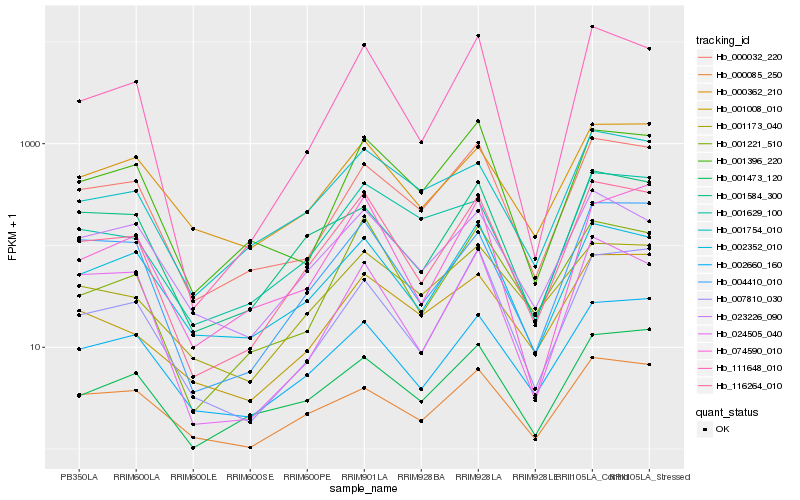
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116264_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 2 |
Hb_111648_010 |
0.0965646044 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 3 |
Hb_001221_510 |
0.1065257129 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 4 |
Hb_004410_010 |
0.108881804 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 5 |
Hb_002660_160 |
0.1097389058 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000032_220 |
0.1137020735 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002352_010 |
0.1188626876 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 8 |
Hb_001584_300 |
0.1214492443 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 9 |
Hb_001473_120 |
0.1238863112 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000085_250 |
0.124332659 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 11 |
Hb_001754_010 |
0.125844365 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 12 |
Hb_074590_010 |
0.1285224893 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 13 |
Hb_000362_210 |
0.129001203 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 14 |
Hb_007810_030 |
0.1300627737 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 15 |
Hb_023226_090 |
0.1336544689 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 16 |
Hb_001396_220 |
0.1337076589 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 17 |
Hb_001629_100 |
0.1366366832 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X2 [Vitis vinifera] |
| 18 |
Hb_001008_010 |
0.136852098 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_001173_040 |
0.1386654139 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_024505_040 |
0.1389176028 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |