Hb_001396_220
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig1396: 159073-161044 |
| Sequence |   |
Annotation
kegg
| ID | rcu:RCOM_1079720 |
|---|---|
| description | nuclear transport factor, putative |
nr
| ID | XP_002514900.1 |
|---|---|
| description | nuclear transport factor, putative [Ricinus communis] |
swissprot
| ID | Q9C7F5 |
|---|---|
| description | Nuclear transport factor 2 OS=Arabidopsis thaliana GN=NTF2 PE=1 SV=1 |
trembl
| ID | B9RMD1 |
|---|---|
| description | Nuclear transport factor, putative OS=Ricinus communis GN=RCOM_1079720 PE=4 SV=1 |
Gene Ontology
| ID | GO:0005635 |
|---|---|
| description | nuclear transport factor 2 |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_10761: 159123-161042 , PASA_asmbl_10763: 159123-159831 |
|---|---|
| cDNA (Sanger) (ID:Location) |
001_P19.ab1: 159175-161028 , 003_L17.ab1: 159174-161028 , 004_O15.ab1: 159227-161028 , 007_E22.ab1: 160365-161026 , 009_M09.ab1: 159387-161022 , 011_M06.ab1: 159182-161026 , 011_N23.ab1: 159125-161033 , 012_F24.ab1: 159180-161033 , 013_L01.ab1: 159236-161027 , 021_A04.ab1: 159125-161033 , 021_E09.ab1: 159246-161033 , 021_L17.ab1: 159177-161020 , 023_D01.ab1: 159386-161026 , 025_K01.ab1: 159184-161033 , 026_O21.ab1: 159310-161028 , 027_L16.ab1: 160365-161026 , 029_O11.ab1: 159362-161027 , 033_P10.ab1: 159252-161033 , 033_P16.ab1: 159174-161031 , 034_E24.ab1: 159184-159390 , 036_J03.ab1: 159184-161032 , 036_K03.ab1: 159185-161023 , 038_J12.ab1: 159123-161033 , 039_G06.ab1: 159185-161033 , 042_H19.ab1: 159233-161027 , 045_B11.ab1: 159185-161023 , 045_M23.ab1: 159185-161033 , 046_C14.ab1: 159228-161026 , 048_J15.ab1: 159263-161033 , 050_E18.ab1: 159220-161020 , 051_G10.ab1: 159291-161033 |
Similar expressed genes (Top20)
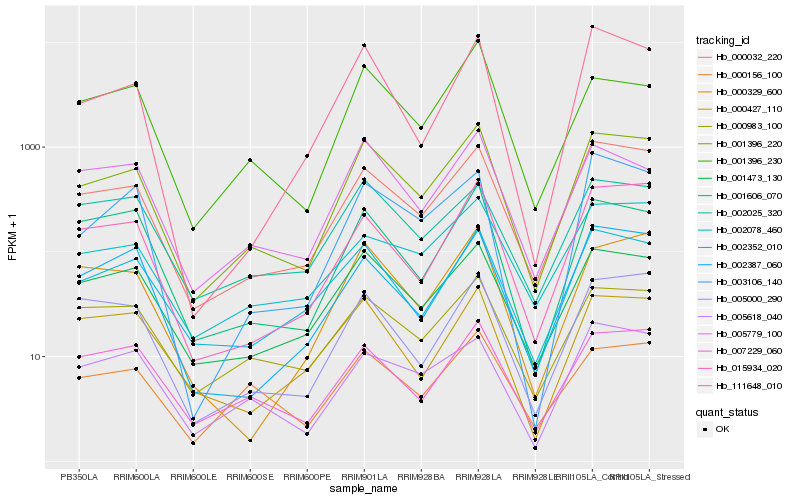
Gene co-expression network
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 622.331 | 110.624 | 32.4376 | 64.7766 | 420.422 | 1153.82 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 1367.5 | 1199.64 | 1664.83 | 329.632 | 41.0427 |

