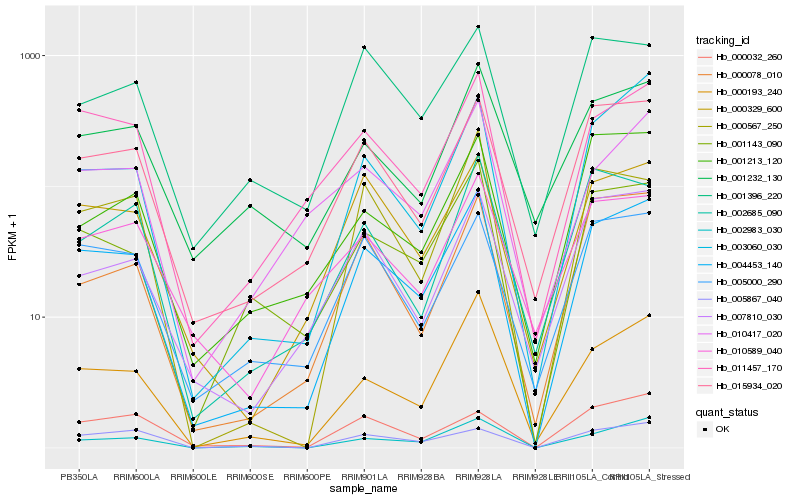
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004453_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104880181 [Vitis vinifera] |
| 2 |
Hb_002983_030 |
0.1084988375 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g046124mg, partial [Citrus sinensis] |
| 3 |
Hb_015934_020 |
0.1101882561 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 4 |
Hb_000193_240 |
0.1128040599 |
- |
- |
hypothetical protein CISIN_1g0449881mg, partial [Citrus sinensis] |
| 5 |
Hb_000329_600 |
0.1182662223 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |
| 6 |
Hb_001143_090 |
0.1204740171 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 6 [Jatropha curcas] |
| 7 |
Hb_011457_170 |
0.1232702237 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 8 |
Hb_003060_030 |
0.1272189467 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 9 |
Hb_005000_290 |
0.1316653738 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 10 |
Hb_000078_010 |
0.1351673899 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 11 |
Hb_000567_250 |
0.1383477365 |
- |
- |
hypothetical protein B456_009G162000 [Gossypium raimondii] |
| 12 |
Hb_007810_030 |
0.145075206 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 13 |
Hb_005867_040 |
0.1496098568 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein RKD4 [Jatropha curcas] |
| 14 |
Hb_001232_130 |
0.1513336226 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 15 |
Hb_010417_020 |
0.1529318047 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 16 |
Hb_000032_260 |
0.1532256582 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 17 |
Hb_002685_090 |
0.1553463162 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 18 |
Hb_001213_120 |
0.1560284455 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 19 |
Hb_001396_220 |
0.1562093186 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 20 |
Hb_010589_040 |
0.1573061798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |