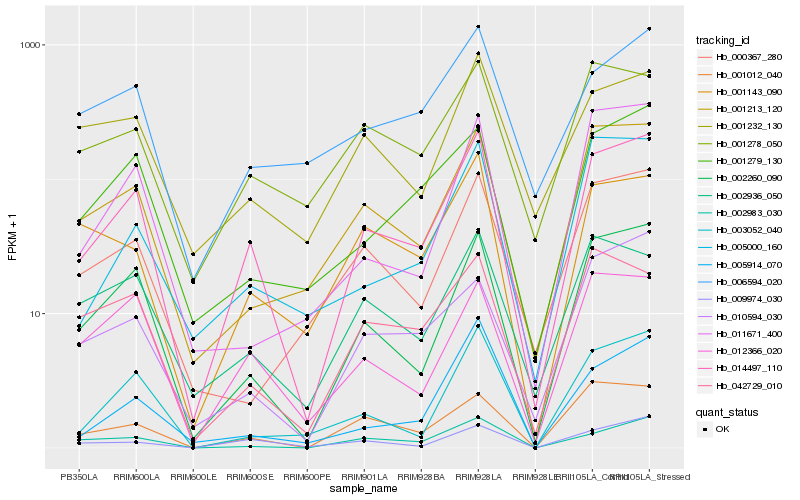
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_110 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 2 |
Hb_002260_090 |
0.106381176 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001213_120 |
0.141504932 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 4 |
Hb_012366_020 |
0.1467240684 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001012_040 |
0.1468746195 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002983_030 |
0.1496327448 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g046124mg, partial [Citrus sinensis] |
| 7 |
Hb_003052_040 |
0.1547690368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001279_130 |
0.1582281999 |
- |
- |
hypothetical protein L484_017676 [Morus notabilis] |
| 9 |
Hb_009974_030 |
0.1605378654 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_002936_050 |
0.1655607322 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001278_050 |
0.1670977709 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 12 |
Hb_005000_160 |
0.1694807732 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 13 |
Hb_042729_010 |
0.1697775478 |
- |
- |
- |
| 14 |
Hb_001143_090 |
0.1701342177 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 6 [Jatropha curcas] |
| 15 |
Hb_000367_280 |
0.1721696312 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 16 |
Hb_010594_030 |
0.1724912921 |
- |
- |
- |
| 17 |
Hb_006594_020 |
0.1726346939 |
- |
- |
PREDICTED: uncharacterized protein LOC105138974 isoform X1 [Populus euphratica] |
| 18 |
Hb_001232_130 |
0.1733982117 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 19 |
Hb_011671_400 |
0.1764046194 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 20 |
Hb_005914_070 |
0.1771127488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |