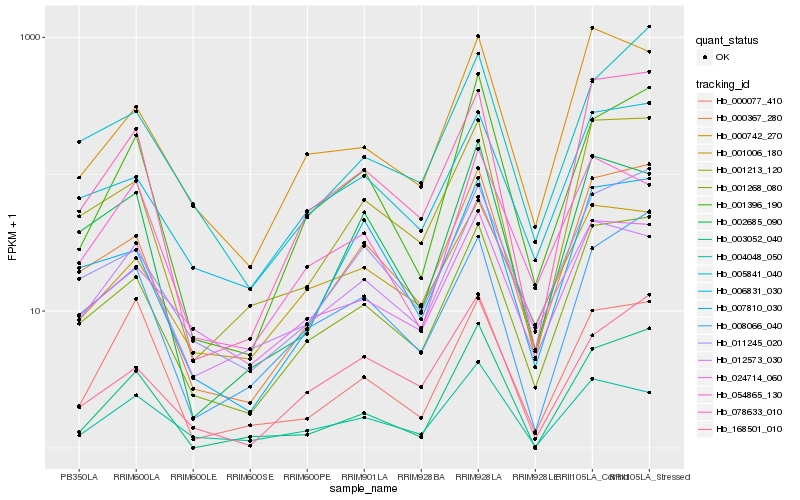
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105123025 [Populus euphratica] |
| 2 |
Hb_003052_040 |
0.127910068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000367_280 |
0.1331768237 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 4 |
Hb_168501_010 |
0.1349635911 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001268_080 |
0.1360460845 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_078633_010 |
0.1546964843 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_008066_040 |
0.1599103106 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL62 [Jatropha curcas] |
| 8 |
Hb_011245_020 |
0.160840934 |
- |
- |
Ferrochelatase [Medicago truncatula] |
| 9 |
Hb_004048_050 |
0.1656951934 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 10 |
Hb_007810_030 |
0.1667263382 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 11 |
Hb_001213_120 |
0.1673867776 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 12 |
Hb_000077_410 |
0.1678109763 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_012573_030 |
0.1731221813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002685_090 |
0.1732316974 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 15 |
Hb_054865_130 |
0.1762938711 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 16 |
Hb_005841_040 |
0.179023317 |
- |
- |
stress-induced hydrophobic peptide 1 [Hevea brasiliensis] |
| 17 |
Hb_024714_060 |
0.1808665094 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006831_030 |
0.1814337184 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 19 |
Hb_001006_180 |
0.18239586 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 20 |
Hb_000742_270 |
0.1827825807 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |