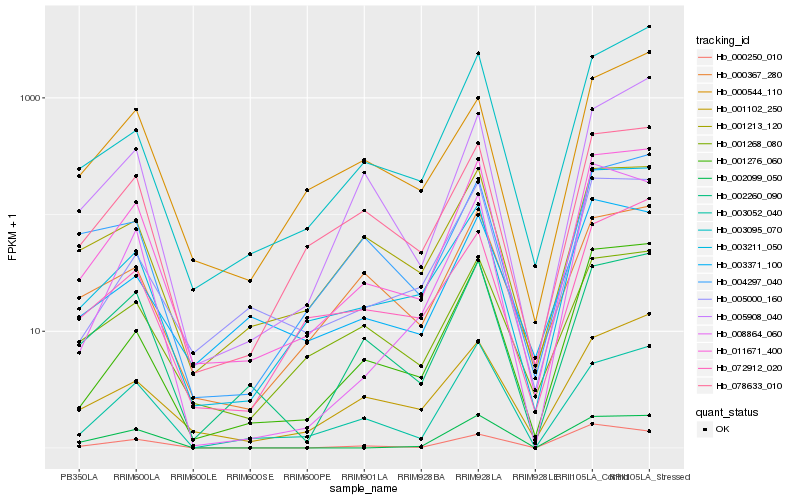
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_400 |
0.0 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 2 |
Hb_001276_060 |
0.1051062283 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 3 |
Hb_078633_010 |
0.1157047947 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 4 |
Hb_002260_090 |
0.1198360073 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003095_070 |
0.1198866477 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 6 |
Hb_003052_040 |
0.1208257449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005000_160 |
0.1219382631 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 8 |
Hb_003211_050 |
0.1242469073 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 9 |
Hb_001102_250 |
0.1281067788 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_001213_120 |
0.1355345974 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 11 |
Hb_005908_040 |
0.1355550506 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 12 |
Hb_072912_020 |
0.1378161414 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 13 |
Hb_001268_080 |
0.1390784922 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000250_010 |
0.1434009084 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 15 |
Hb_004297_040 |
0.1467995369 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_000367_280 |
0.1472653621 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 17 |
Hb_003371_100 |
0.1474455218 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_000544_110 |
0.1474603313 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 19 |
Hb_008864_060 |
0.1490787614 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 20 |
Hb_002099_050 |
0.1497986933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |