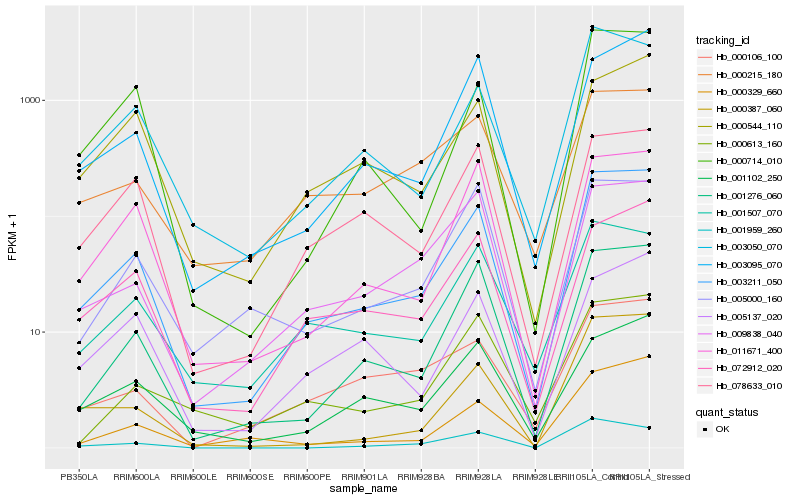
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003211_050 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 2 |
Hb_001276_060 |
0.1076880201 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 3 |
Hb_003050_070 |
0.1185908945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011671_400 |
0.1242469073 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 5 |
Hb_072912_020 |
0.125184648 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_000714_010 |
0.1291434957 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 7 |
Hb_003095_070 |
0.1305357167 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 8 |
Hb_009838_040 |
0.1356523252 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 9 |
Hb_001507_070 |
0.1360021767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001102_250 |
0.1382058925 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_078633_010 |
0.138722752 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 12 |
Hb_000387_060 |
0.1388022468 |
- |
- |
- |
| 13 |
Hb_000329_660 |
0.1392212543 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000544_110 |
0.1430541679 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 15 |
Hb_005000_160 |
0.1470804275 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 16 |
Hb_001959_260 |
0.1475527902 |
- |
- |
- |
| 17 |
Hb_000215_180 |
0.1508327709 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 18 |
Hb_005137_020 |
0.1508862548 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 19 |
Hb_000106_100 |
0.1516741794 |
- |
- |
- |
| 20 |
Hb_000613_160 |
0.154841267 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |