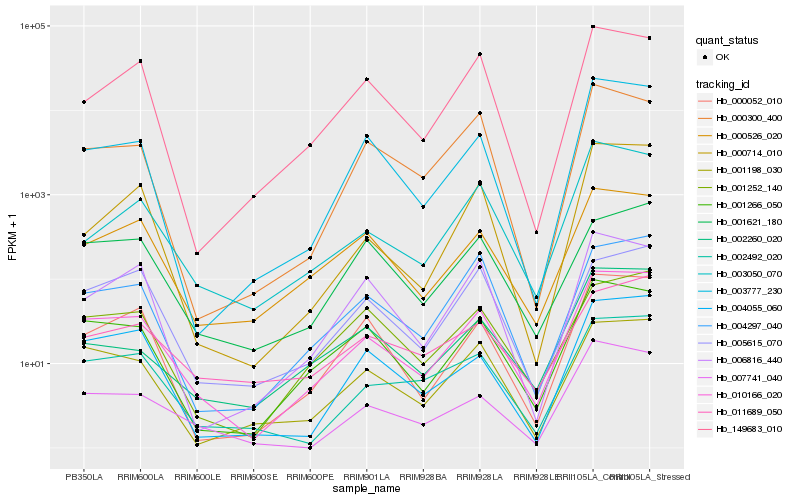
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010166_020 |
0.0 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001266_050 |
0.1072126635 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 3 |
Hb_002492_020 |
0.1124877142 |
- |
- |
PREDICTED: uncharacterized protein LOC104789613 [Camelina sativa] |
| 4 |
Hb_001198_030 |
0.1189435313 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 5 |
Hb_000052_010 |
0.1194932136 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 6 |
Hb_000526_020 |
0.1239067427 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 7 |
Hb_007741_040 |
0.1240949216 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105643153 [Jatropha curcas] |
| 8 |
Hb_004055_060 |
0.1243753228 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 9 |
Hb_003777_230 |
0.1248339797 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 10 |
Hb_004297_040 |
0.1340967983 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 11 |
Hb_003050_070 |
0.1369397813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001252_140 |
0.1393772083 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_149683_010 |
0.1404972562 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 14 |
Hb_000300_400 |
0.1450577818 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 15 |
Hb_000714_010 |
0.1454019377 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 16 |
Hb_002260_020 |
0.1459962201 |
- |
- |
- |
| 17 |
Hb_011689_050 |
0.1494902401 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 18 |
Hb_006816_440 |
0.1525002905 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_005615_070 |
0.1525736741 |
- |
- |
Rab7 [Hevea brasiliensis] |
| 20 |
Hb_001621_180 |
0.1551845221 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |