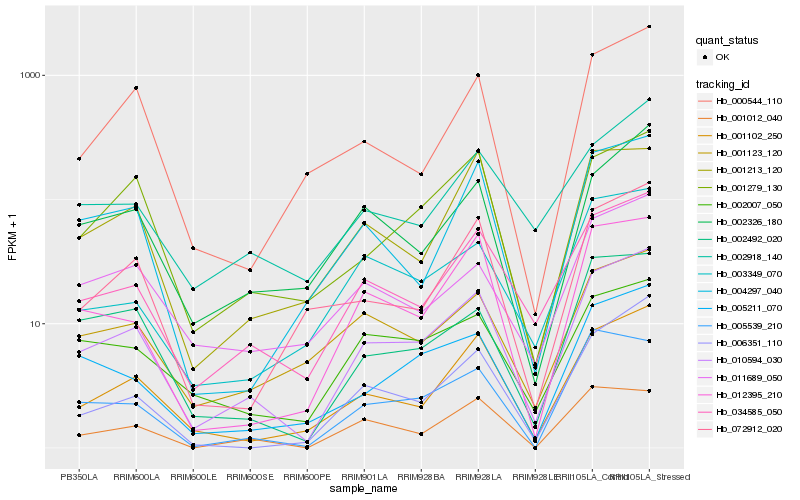
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010594_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_034585_050 |
0.1029704893 |
- |
- |
- |
| 3 |
Hb_001102_250 |
0.1104644118 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_002492_020 |
0.1283337382 |
- |
- |
PREDICTED: uncharacterized protein LOC104789613 [Camelina sativa] |
| 5 |
Hb_005211_070 |
0.1294567696 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 6 |
Hb_012395_210 |
0.1338215556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006351_110 |
0.1359449373 |
- |
- |
- |
| 8 |
Hb_002326_180 |
0.1377970353 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 9 |
Hb_001279_130 |
0.1387262686 |
- |
- |
hypothetical protein L484_017676 [Morus notabilis] |
| 10 |
Hb_005539_210 |
0.1401956269 |
- |
- |
- |
| 11 |
Hb_001123_120 |
0.1421734461 |
- |
- |
Uncharacterized protein TCM_042776 [Theobroma cacao] |
| 12 |
Hb_004297_040 |
0.1426768355 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 13 |
Hb_011689_050 |
0.144448095 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 14 |
Hb_003349_070 |
0.1453607338 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 15 |
Hb_002918_140 |
0.1457981295 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 16 |
Hb_072912_020 |
0.1481888507 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 17 |
Hb_001213_120 |
0.148552585 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 18 |
Hb_000544_110 |
0.1486235122 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 19 |
Hb_002007_050 |
0.1488333623 |
- |
- |
PREDICTED: putative protease Do-like 14 [Jatropha curcas] |
| 20 |
Hb_001012_040 |
0.1507679396 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |