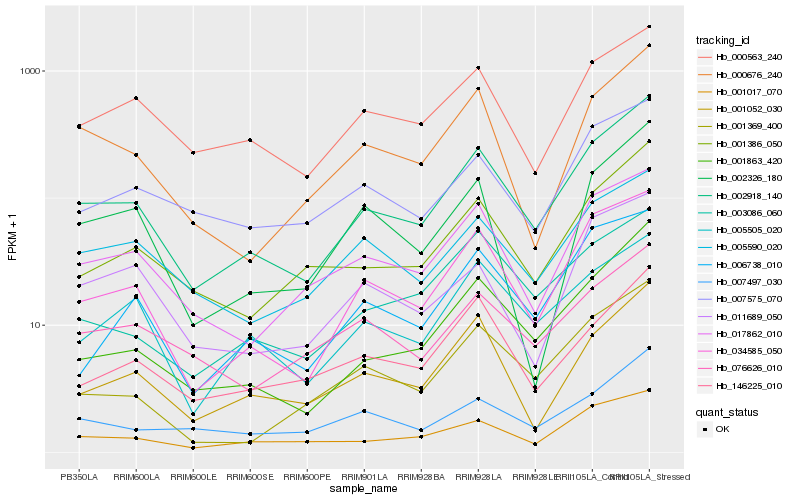
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_140 |
0.0 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 2 |
Hb_034585_050 |
0.0849854945 |
- |
- |
- |
| 3 |
Hb_001863_420 |
0.096072371 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 4 |
Hb_001386_050 |
0.1049635235 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_001017_070 |
0.1078890656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_017862_010 |
0.1135232882 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 7 |
Hb_000676_240 |
0.1170385127 |
- |
- |
thioredoxin H-type 2 [Hevea brasiliensis] |
| 8 |
Hb_001052_030 |
0.120338568 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000563_240 |
0.1221563885 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 10 |
Hb_005505_020 |
0.1239776259 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |
| 11 |
Hb_001369_400 |
0.1258540082 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 12 |
Hb_007497_030 |
0.1275901633 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 13 |
Hb_146225_010 |
0.1277495353 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 14 |
Hb_003086_060 |
0.1292392867 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 15 |
Hb_005590_020 |
0.1304115386 |
- |
- |
hypothetical protein CICLE_v10010008mg [Citrus clementina] |
| 16 |
Hb_076626_010 |
0.1337190623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007575_070 |
0.1338745746 |
- |
- |
Histone H3.2 [Glycine soja] |
| 18 |
Hb_011689_050 |
0.134873143 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 19 |
Hb_006738_010 |
0.1349292784 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 20 |
Hb_002326_180 |
0.1363091327 |
- |
- |
Protein Z, putative [Ricinus communis] |