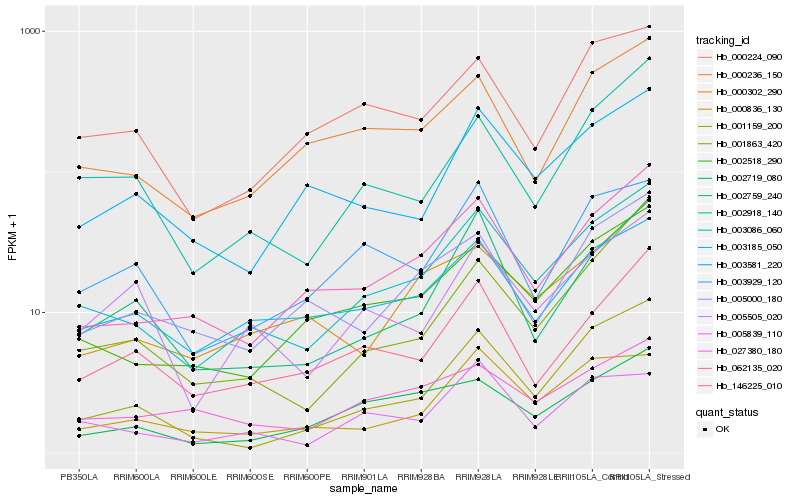
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 2 |
Hb_002518_290 |
0.0982052039 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 3 |
Hb_062135_020 |
0.1082354329 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 4 |
Hb_003185_050 |
0.1227191722 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 5 |
Hb_000236_150 |
0.1260588583 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_001159_200 |
0.128327735 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002918_140 |
0.1292392867 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 8 |
Hb_000836_130 |
0.1318955297 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002759_240 |
0.1332309381 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 10 |
Hb_000302_290 |
0.1345177256 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 11 |
Hb_002719_080 |
0.1347148217 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 12 |
Hb_005839_110 |
0.1354481414 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |
| 13 |
Hb_000224_090 |
0.1354516672 |
- |
- |
Eukaryotic translation initiation factor 1A [Gossypium arboreum] |
| 14 |
Hb_146225_010 |
0.1361370011 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 15 |
Hb_003929_120 |
0.1379980568 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_027380_180 |
0.1383170098 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 17 |
Hb_005000_180 |
0.1399731739 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 18 |
Hb_001863_420 |
0.1403617596 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 19 |
Hb_003581_220 |
0.1409406497 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_005505_020 |
0.1415943042 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |