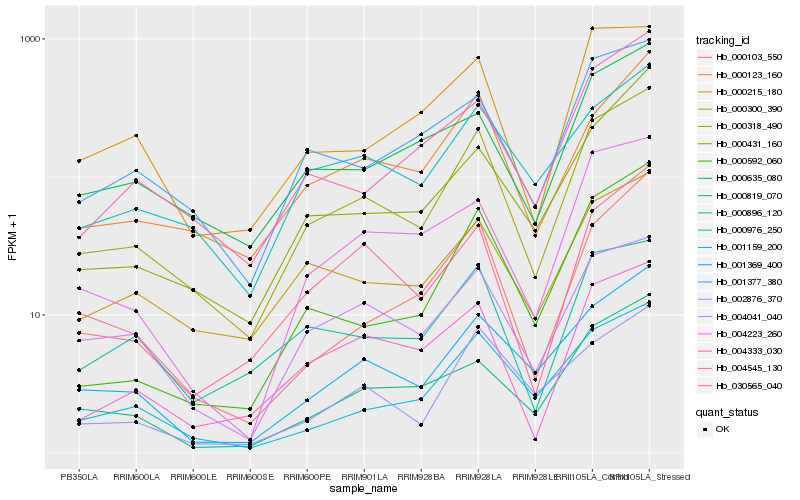
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_490 |
0.0 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 2 |
Hb_000123_160 |
0.1036479556 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 3 |
Hb_001369_400 |
0.1124417899 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 4 |
Hb_030565_040 |
0.1130400979 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_000976_250 |
0.1152382788 |
- |
- |
- |
| 6 |
Hb_004333_030 |
0.115863068 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 7 |
Hb_001377_380 |
0.1163890741 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 8 |
Hb_000300_390 |
0.1172825734 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 9 |
Hb_004545_130 |
0.1197084496 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 10 |
Hb_000103_550 |
0.1206518796 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001159_200 |
0.1226996143 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000635_080 |
0.1240942844 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 13 |
Hb_004041_040 |
0.1247960192 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM14-1 [Camelina sativa] |
| 14 |
Hb_000819_070 |
0.1256976706 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 15 |
Hb_000431_160 |
0.1258045765 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 16 |
Hb_004223_260 |
0.1276416423 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 17 |
Hb_000592_060 |
0.1287792472 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 18 |
Hb_000896_120 |
0.1295263511 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 19 |
Hb_002876_370 |
0.1299789803 |
- |
- |
hypothetical protein B456_013G140400 [Gossypium raimondii] |
| 20 |
Hb_000215_180 |
0.1309737146 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |