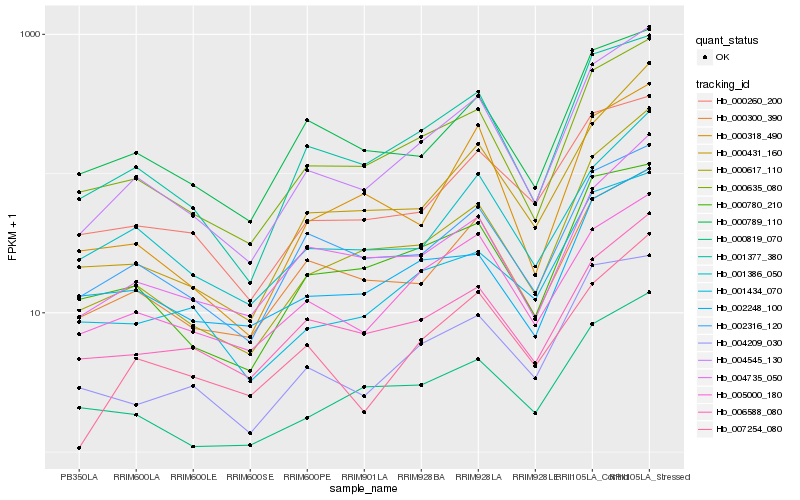
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_130 |
0.0 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 2 |
Hb_000635_080 |
0.0786605297 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 3 |
Hb_001377_380 |
0.080884592 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 4 |
Hb_000617_110 |
0.0857217919 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 5 |
Hb_000431_160 |
0.0983965513 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 6 |
Hb_004735_050 |
0.1076330156 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004209_030 |
0.1111812791 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006588_080 |
0.1113619835 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 9 |
Hb_001434_070 |
0.1120112928 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 10 |
Hb_002316_120 |
0.1153268342 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 11 |
Hb_000780_210 |
0.1170416546 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 12 |
Hb_001386_050 |
0.117962472 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 13 |
Hb_000300_390 |
0.1194728507 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 14 |
Hb_000318_490 |
0.1197084496 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 15 |
Hb_007254_080 |
0.12245108 |
- |
- |
- |
| 16 |
Hb_000789_110 |
0.1234325793 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 17 |
Hb_000819_070 |
0.1238531239 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 18 |
Hb_005000_180 |
0.124936263 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 19 |
Hb_000260_200 |
0.1258083126 |
- |
- |
- |
| 20 |
Hb_002248_100 |
0.1284313413 |
- |
- |
hypothetical protein VITISV_027197 [Vitis vinifera] |