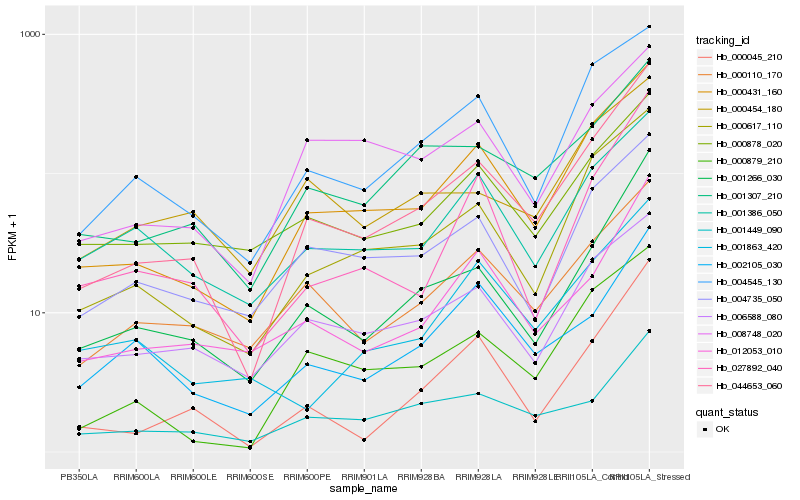
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044653_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 2 |
Hb_000431_160 |
0.0753166663 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 3 |
Hb_001266_030 |
0.0941008626 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000617_110 |
0.1061330541 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 5 |
Hb_000045_210 |
0.12204481 |
- |
- |
- |
| 6 |
Hb_000879_210 |
0.1254801639 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 7 |
Hb_004545_130 |
0.1293798853 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 8 |
Hb_012053_010 |
0.1309706381 |
- |
- |
srpk, putative [Ricinus communis] |
| 9 |
Hb_001307_210 |
0.1331628233 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 10 |
Hb_027892_040 |
0.1352723546 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 11 |
Hb_000110_170 |
0.1432226187 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_004735_050 |
0.144525397 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001449_090 |
0.1497489394 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006588_080 |
0.1557260532 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 15 |
Hb_000878_020 |
0.1558632486 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 16 |
Hb_002105_030 |
0.1587491834 |
- |
- |
- |
| 17 |
Hb_008748_020 |
0.1610565541 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 18 |
Hb_001386_050 |
0.16126532 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 19 |
Hb_000454_180 |
0.1626455571 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 20 |
Hb_001863_420 |
0.1644174834 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |