| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
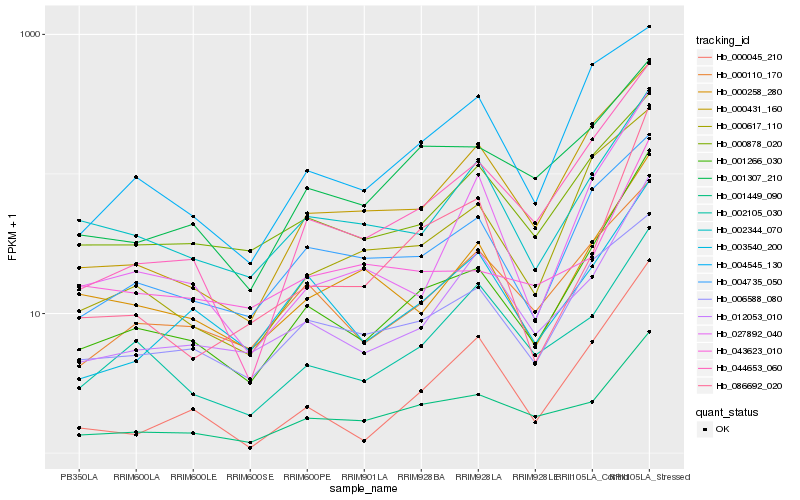
Hb_001266_030 |
0.0 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_044653_060 |
0.0941008626 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 3 |
Hb_012053_010 |
0.120783975 |
- |
- |
srpk, putative [Ricinus communis] |
| 4 |
Hb_000045_210 |
0.1331068793 |
- |
- |
- |
| 5 |
Hb_027892_040 |
0.1345136682 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 6 |
Hb_000431_160 |
0.1347055045 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 7 |
Hb_000617_110 |
0.1435690051 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 8 |
Hb_086692_020 |
0.1526236807 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_004735_050 |
0.1527255662 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000258_280 |
0.1538845396 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 11 |
Hb_001449_090 |
0.15764071 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002344_070 |
0.1597942334 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 13 |
Hb_001307_210 |
0.1612488423 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 14 |
Hb_000110_170 |
0.1622273061 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003540_200 |
0.1626879214 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 16 |
Hb_000878_020 |
0.164320725 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 17 |
Hb_043623_010 |
0.1676524438 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 18 |
Hb_002105_030 |
0.168189173 |
- |
- |
- |
| 19 |
Hb_004545_130 |
0.1682348688 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 20 |
Hb_006588_080 |
0.1717220428 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |