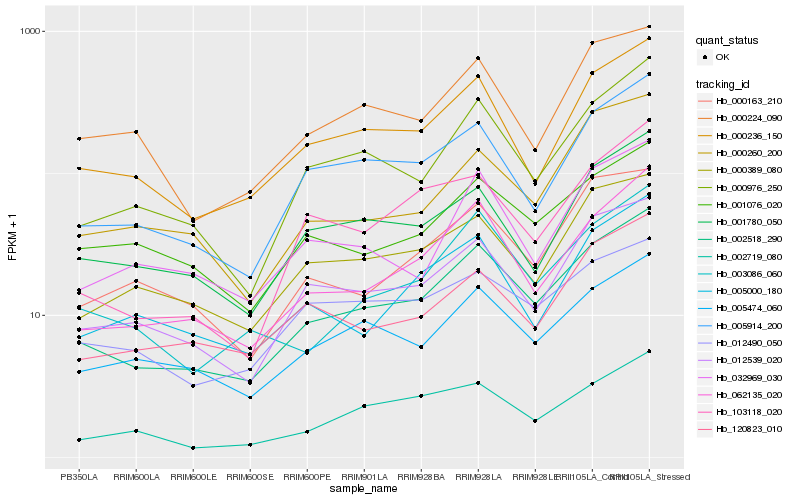
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002518_290 |
0.0 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 2 |
Hb_062135_020 |
0.0769880149 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 3 |
Hb_000976_250 |
0.0850390344 |
- |
- |
- |
| 4 |
Hb_012539_020 |
0.0889790737 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 5 |
Hb_005914_200 |
0.0890587268 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 6 |
Hb_000236_150 |
0.0891124228 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_003086_060 |
0.0982052039 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 8 |
Hb_005474_060 |
0.1003963262 |
- |
- |
hypothetical protein PHAVU_011G019700g [Phaseolus vulgaris] |
| 9 |
Hb_002719_080 |
0.1059371117 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 10 |
Hb_001780_050 |
0.1067890477 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC2 [Jatropha curcas] |
| 11 |
Hb_000389_080 |
0.1090042861 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 12 |
Hb_103118_020 |
0.1101290581 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 13 |
Hb_000163_210 |
0.1115716153 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 14 |
Hb_120823_010 |
0.1118698742 |
- |
- |
- |
| 15 |
Hb_000224_090 |
0.1141499078 |
- |
- |
Eukaryotic translation initiation factor 1A [Gossypium arboreum] |
| 16 |
Hb_000260_200 |
0.1146868728 |
- |
- |
- |
| 17 |
Hb_005000_180 |
0.1151740847 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 18 |
Hb_001076_020 |
0.1152442471 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 19 |
Hb_032969_030 |
0.119254145 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_012490_050 |
0.1200151219 |
- |
- |
PREDICTED: uncharacterized protein LOC105637921 [Jatropha curcas] |